
Research Article
Austin J Pulm Respir Med. 2021; 8(1): 1070.
Prediction Model of Acute Respiratory Distress Syndrome for Hospitalized Patients with Covid-19 Pneumonia
Zhang J1#, Liu X2#, Yang J2#, Wang F1, Yang C1, Jiang X1, Su L1* and Peng Z1*
¹Department of Critical Care Medicine, Zhongnan Hospital of Wuhan University, China
²Department of Respiratory and Critical Care Medicine, Wuhan Fourth Hospital, China
#Contributed equally to this study
*Corresponding author: Zhiyong Peng, Department of Critical Care Medicine, Zhongnan Hospital of Wuhan University, Hubei 430000, China. Email: pengzy5@ hotmail.com
Lianjiu Su, Department of Critical Care Medicine, Zhongnan Hospital of Wuhan University, Hubei 430000, China. Email: sulianjiu@whu.edu.cn
Received: April 03, 2021; Accepted: April 22, 2021; Published: April 29, 2021
Abstract
Background: COVID-19 pneumonia has become a worldwide epidemic. Acute Respiratory Distress Syndrome (ARDS) is a major cause of mortality. Early recognition the risk of ARDS of COVID-19 patients is vital.
Methods: Descriptive study from Zhongnan Hospital of Wuhan University and Wuhan Fourth Hospital. 394 consecutive hospitalized patients with confirmed COVID-19 infection from January 1 to March 15, 2020.
Results: We developed a risk prediction model of ARDS for COVID-19 among 394 enrolled patients. The variables included in the model were sex, age, diabetes mellitus, neutrophil and lymphocyte counts, serum urea levels, and pulmonary lesion range. The model performed well in predicting ARDS occurrence with excellent discrimination (C-stat=0.81) and appropriate calibration. The predictive value of our model was better than that of the Lung Injury Prediction Score (LIPS) in the discovery set [AUC: 0.77 (0.71, 0.82) vs 0.68 (0.61, 0.75), P=0.02].
Conclusions: Our prediction model provides clinicians and researchers a simple tool to screen for COVID-19 patients at high risk of ARDS. Potential clinical benefits of using this model deserve assessment.
Keywords: Pneumonia; COVID-19; Early prediction; Model; ARDS
Introduction
In December 2019, a cluster of acute respiratory illnesses, now known as COVID-19, occurred in Wuhan, Hubei Province, China [1]. In recent days, infections have rapidly spread from Wuhan to other areas and in more than 190 countries around the world [2,3]. Several studies have reported the epidemiological and clinical characteristics of COVID-19 with a mortality of 4.3% in Wuhan [3- 5]. Acute Respiratory Distress Syndrome (ARDS) is a primary cause of death in many COVID-19 patients. It was reported that more than 60% of COVID-19 patients in intensive care units developed ARDS and most of the patients eventually died of severe ARDS [6]. Our previous study has confirmed that the median time from the first symptom to ARDS is about eight days [6]. Thus, it is essential to recognize the ARDS risk factors early and prevent its development or progression in COVID-19 patients. Previous studies showed that the Lung Injury Prediction Score (LIPS) could predict ARDS early, which included four indicators, such as susceptibility factors, high-risk surgery or trauma, interventions to mitigate the risk [7]. However, this scoring model might not fit in COVID-19 patients due to different study cohort with different etiology and pathogenesis [8]. A proprietary prediction model is urgently needed to identify the risk of ARDS in COVID-19 patients, which may be provide the chance to implement effective preventive strategies to improve patients’ clinical outcome. The aim of the current study was to develop and validate a model to predict the risk of ARDS in COVID-19 patients.
Methods
Study Population
Retrospective data from two centers, i.e., Zhongnan Hospital of Wuhan University and Wuhan Fourth Hospital, Wuhan city in China between January 1 to March 15, 2020, were utilized. We included all consecutive patients with COVID-19 who were diagnosed according to World Health Organization interim guidance in this study [4]. Reverse Transcription-Polymerase Chain Reaction (RT-PCR) was used as a gold standard to diagnose COVID-19 in multiple and different clinical specimens when necessary. We excluded patients who presented ARDS on admission. The ethical committees of Wuhan University and Wuhan Forth hospital approved the project (No. 2020020 and No.202002, respectively), and we obtained oral consent from patients or patients’ relatives.
Data collection
Epidemiological, clinical, laboratory, and radiological characteristics and treatment and outcomes data were abstracted using electronic medical records. The information recorded included demographics, comorbidities, laboratory findings (blood routine, liver function, renal function,) and LIPS calculated on the first admission day, imaging data (chest CT scans, and X-rays) during the first two admission days. The investigators followed the same protocols and definitions to review and analyze the collected data and were blinded to the patients’ ARDS status. A consensus resolved any disagreements between the investigators.
Outcome variables
The primary outcome was the development of ARDS during the hospitalization. ARDS diagnosis was according to the Berlin definition established in 2012 [9], including the development of bilateral pulmonary infiltrates on chest radiography, PaO2/FiO2 ratio <300, and the absence of left atrial hypertension as the primary explanation for pulmonary edema. The diagnosis of ARDS was made by consensus among two physicians.
Sample size estimation
The available sample size was evaluated to ensure there would be an adequate number of ARDS cases to support logistic regression modeling. The previous study suggested that the ARDS incidence in this cohort would be 19.6% [6]. Using the standard rule of at least ten events for each covariate, our study sample was believed to be sufficiently large.
Statistical analysis, model establishment, and validation
Continuous characteristics were reported by median (interquartile range), categorical variables by number (%). Differences between the two groups were compared with the t-test, chi-square test, or Mann Whitney U test accordingly. Missing data occurred only in liver and kidney function tests, accounting for <2% due to the absence of the test results. We used the multiple imputation method to impute the missing data. Categorical variables were transformed into dummy variables for analysis. The Least Absolute Shrinkage and Selection Operator (LASSO) method, suitable for the reduction in high dimensional data, was used to regularize a logistic regression model to build the prediction model [10]. For model training and validation, the entire cohort was randomly divided into discovery and validation sets using a 7:3 ratio. Optimal parameter (lambda) giving the most regularized model was tuned through 10-fold cross-validation using the R package “glmnet” (version 2.0-16) [11,12]. Receiver Operating Characteristic (ROC) curves were generated to evaluate the LASSO-regularized logistic regression model in the discovery set and validation set. Features selecting through the LASSO were incorporated into a multivariable logistic regression model to build a nomogram. The nomogram was internally validated through 1000 bootstrap resamples. Calibration analysis [13] were performed to evaluate the calibration. The corresponding nomogram and calibration curve were drawn using the R package “rms” (version 5.1- 3). Analyses were performed using SPSS 22.0 software and R version 3.5.2. Any difference at P-value<0.05 was considered statistically significant.
Results
Patient characteristics
Of the 394 patients with COVID-19 infection included in our study, 117 (29.7%) patients developed ARDS, among them, 22(18.8%) patients died. The median time from admission to the ARDS diagnosis was 4 (2-6) days. The median age of those with and without ARDS was statistically different, i.e., 61 years old for ASRDS group and 52 years old non-ARDS patients (p<0.05). Patients with hypertension, diabetes mellitus, cerebrovascular disease were more likely to develop ARDS (p<0.05). Lymphocyte and platelet counts, and albumin in the ARDS group were significantly lower than that non-ARDS group. Neutrophil count, alanine aminotransferase, and aspartate aminotransferase in the ARDS group were substantially higher than those in the non-ARDS group. The range of lung lesions and their bilateral distribution was significantly higher in the ARDS group. The median length of hospital stay in the ARDS group was significantly higher than that in the non-ARDS group (Table 1).
Factors
All (n=394)
ARDS (n=117)
N-ARDS (n=277)
P value
Age (IQR)
56 (42-67)
61 (52-73)
53 (39-63)
<0.001
Sex n (%)
Male
186 (47.2)
66 (56.4)
120 (43.3)
0.017
Female
208 (52.8)
51 (43.6)
157 (56.7)
Comorbidities n (%)
Hypertension
115 (29.2)
47 (40.2)
68 (24.5)
0.002
Diabetes n
47 (11.9)
25 (21.4)
22 (7.9)
<0.001
Cardiovascular disease
38 (9.6)
15 (12.8)
23 (8.3)
0.165
Cerebrovascular disease
15 (3.8)
9 (7.7)
6 (2.2)
0.009
Chronic pulmonary disease
23 (5.8)
7 (6.0)
16 (5.8)
0.936
Chronic kidney disease
4 (1.0)
2 (1.7)
2 (0.7)
0.372
Malignancy
24 (6.1)
11 (9.4)
13 (4.7)
0.074
Temperature (IQR)
37.6 (36.7-38.5)
37.9 (36.6-38.7)
37.6 (36.8-38.4)
0.793
Hear rate (IQR)
85 (78-95)
88 (80-97)
85 (78-93)
0.109
Respiratory rate (IQR)
20 (18-21)
20 (18-22)
20 (18-21)
0.43
Blood pressure (IQR)
Systolic pressure
125 (118-135)
130 (120-137)
124 (117-134)
0.03
Diastolic pressure
76 (70-82)
75 (70-84)
77 (70-82)
0.462
Mean arterial pressure
93.3 (86.6-98.3)
93.30 (86.6-100.0)
93.3 (86.6-98.0)
0.548
Laboratory data
WBC count, ×109/L (IQR)
4.58 (3.38-6.06)
4.94 (3.21-7.14)
4.46 (3.44-5.71)
0.092
Neutrophil count, ×109/L (IQR)
3.12 (2.13-4.67)
3.87 (2.32-6.15)
2.89 (2.01-4.09)
<0.001
Lymphocyte count, ×109/L (IQR)
0.90 (0.62-1.23)
0.70 (0.49-1.05)
0.95(0.68-1.28)
<0.001
Platelet count, ×109/L (IQR)
169.5 (128.3-212.0)
154.0 (122.0-196.8)
174.5 (131.2-219.8)
0.017
Hemoglobin concentration (IQR)
125.7 (118.0-134.0)
123.9 (118.2-135.0)
125.7 (116.0-134.0)
0.998
ALT, U/L (IQR)
25.0 (16.0-41.0)
30.0 (18.5-48.5)
23.0 (15.0-37.8)
0.005
AST, U/L (IQR)
30.0 (23.0-44.5)
38.0 (27.0-60.0)
28.0 (22.0-37.0)
<0.001
Creatinine, μmol/L (IQR)
69.0 (55.7-83.1)
75.1 (61.0-86.3)
67.0 (55.2-79.6)
0.001
Urea, mmol/L (IQR)
4.35 (3.45-5.68)
5.26 (4.09-7.07)
4.01 (3.23-5.21)
<0.001
Radiology
Bilateral distribution n (%)
348 (88.3)
111 (94.9)
237 (85.6)
0.009
Lesion range (>50%) n (%)
57 (14.5)
28 (23.9)
29 (10.5)
0.001
Hydrothorax n (%)
12 (3.0)
5 (4.3)
7 (2.5)
0.357
Outcome
Death n (%)
22 (5.6)
22 (18.8)
0 (0)
<0.001
Hospital stay (IQR)
13 (10-16)
14 (10-17)
12 (9-14)
0.004
LIPS (IQR)
6.00 (5.00-7.00)
6.50 (5.50-8.00)
5.50 (4.50-6.50)
<0.001
Notes: Abbreviations: N-ARDS: Non-ARDS; ALT: alanine aminotransferase; AST: aspartate aminotransferase; Lesion range, a CT scan or chest X-ray showing opacities over 50% of total or not.
Table 1: Demographics and characteristics of COVID-19 patients.
Prognostic factors of ARDS
Twenty-eight variables that were different between the two groups were identified. Then they were reduced to seven potential predictors based on 394 patients in the cohort (Supplementary Figure 1). The final seven variables were analyzed with non-zero coefficients in the LASSO regression model. Meanwhile, 276 and 118 patients categorized into the discovery set and validation sets, respectively. As a result, seven clinical features (sex, age, history of diabetes mellitus, Neutrophil Count (N), Lymphocyte Count (L), blood urea level, and pulmonary lesion range with non-zero coefficients in the model were derived and applied to build the prediction model (Table 2).
Variables
LASSO model
Multivariable logistic regression
Coefficients
OR
LCI
UCI
P value
Age
0.003
1.021
1.003
1.039
0.02
Lymphocyte
-0.242
0.348
0.177
0.647
0.001
Neutrophil
0.048
1.082
0.99
1.187
0.088
Urea
0.022
1.12
1.049
1.222
0.003
Sex
-0.133
1.557
0.954
2.553
0.077
Diabetes
-0.316
2.087
1.036
4.206
0.039
Lesion range
-0.141
2.613
1.39
4.919
0.003
Note: LCI, lower confidence interval; UCI, upper confidence interval.
Table 2: Prediction factors for ARDS in COVID-19.
Performance of nomogram and internal validation
The chosen variables, mentioned above, were included in a multivariable logistic model, which was used to construct a nomogram to facilitate the clinical application of the model (Table 2 and Figure 1A). Internally validation with 1000 bootstrap resamples suggested that the nomogram performed well in clinical settings (corrected C-stat equaled to 0.81). The calibration curve of the nomogram for the prediction of ARDS risk in COVID-19 patients demonstrated good agreement in this cohort (Figure 1B). The P-value of Hosmer-Leme show goodness of fit test was .38.
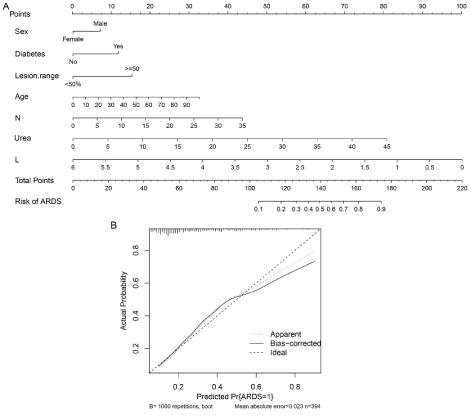
Figure 1: Development and performance of the nomogram.
Notes: A: Nomogram graph of predictive model. To provide a quantitative method to better stratify patients with different classes, a nomogram of ARDS was
constructed integrating significant independent factors identified in the multivariate analysis. B: Calibration curves of the nomogram prediction in the cohort.
Notes: The x-axis represents the predicted ARDS risk. The y-axis represents the actual diagnosed ARDS. The diagonal dotted line represents a perfect prediction
by an ideal model. The solid line represents the performance of the nomogram, of which a closer fit to the diagonal dotted line represents a better prediction.
Abbreviations: N, neutrophil; L, lymphocyte; Lesion range, range of opacity from chest CT or X-ray (opacities over 50% of total or not).
Comparison of the model with LIPS
Using receiver operating characteristic analysis, the predictive value of our model for the incidence of ARDS was compared to that of the LIPS models. The results showed that the performance of our prediction model was good both in the discovery set [AUC=0.77 (0.71, 0.82), specificity=0.57, sensitivity=0.87, Figure 2A] and validation set [AUC=0.81 (0.72, 0.89), specificity=0.78, sensitivity=0.77, Figure 2B]. Our model performed better than LIPS score in the discovery set [AUC: 0.77 (0.71, 0.82) vs 0.68 (0.61, 0.75), P=0.02], however, there was no difference in validation set [AUC: 0.81 (0.72, 0.89) vs 0.73(0.63, 0.73), P= 0.11] (Figure 2).
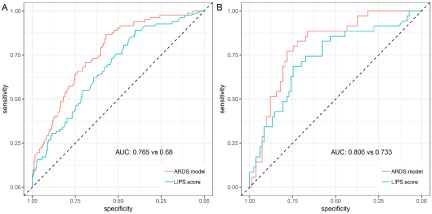
Figure 2: Receiver operating Characteristic (ROC) analysis of the model and LIPS model.
Note: A: Performance in the discovery set; B, Performance in validation set; the red line is our model of AUC and the blue one is LIPS scoring model.
Discussion
In this study of patients with COVID-19, we showed that 29% of patients developed ARDS, and all of the deaths happened in those who developed ARDS. In this multicenter investigation on COVID-19 patients, who were at risk for ARDS development, we tested the predictive accuracy of an ARDS prediction model with good performance. To our knowledge, this was the first prediction model to achieve the goal for ARDS prediction. A nomogram was established and used to estimate the predictive probability of ARDS by collecting readily available variables of sex, age, diabetes mellitus, Neutrophil Count (N), Lymphocyte Count (L), blood urea level, and pulmonary lesion range. For example, a 65-year-old man with diabetes, lymphocyte count of 0.5×109/L, neutrophil count of 3×109/L, blood urea level of 10 mmol/L, and bilateral diffusion in chest CT, could be mapped with the scores of male= 7, age= 20, diabetes mellitus= 12, lymphocyte count= 92, neutrophil count= 2, blood urea level= 15, chest CT= 15; then, all scores were added to give a sum of 163. Finally, we could read the predictive probability corresponding to a score of 163 on the whole score line, which was appropriately 0.8, indicating the probability of ARDS of COVID-19 was 80%. Our results showed that the model had good discrimination with a C-stat of 0.81 along with excellent calibration. However, among patients with an actual probability of >60%, the model might overestimate ARDS risk (Figure 1B). Some individuals with a predicted risk of ARDS of 80% might have an actual probability of approximately 70%. Therefore, the use of the model among those with a very high predicted risk of ARDS should be performed with caution.
Low lymphocyte counts in viral pneumonia is associated with a high mortality rate [14]. In our model, high neutrophils and low lymphocyte counts indicated high levels of inflammation and low levels of immunocompetence. These patients, thus, were predicted to have a high rate of ARDS and worse outcomes following COVID-19. Our results were similar to the previous study that described high neutrophil count, and low lymphocyte count predicted worse outcomes [15]. It has been reported that the incidence of Acute Kidney Injury (AKI) in patients with ARDS could be as high as 35% when patients are admitted to ICU [16]. The mortality rate in patients with ARDS complicated by AKI can reach up to 80% [17]. The elevation of blood urea level indicated azotemia and was possibly associated with clinical or subclinical AKI. In our study, the blood urea levels were monitored in patients with ARDS and we showed similar findings with previous reports. Pulmonary lesion range represented the scale of lung injury, which influenced the pulmonary gas exchange capacity [18]. Chest CT is an important method for diagnosing ARDS [19], and pulmonary lesion range data could be easily obtained from chest CT computer accurately. However, when only the standard chest X-ray was available, the scientists could also evaluate the pulmonary lesion range by estimates when the infiltrations are present in >50% of lung fields. Previous studies showed that diabetic patients had a lower risk of ARDS [20], which might be due to abnormal neutrophil function in diabetic patients. These abnormalities in neutrophil function in diabetic patients may protect the lung by decreasing the ability of these cells to migrate and damage the lungs by oxidative stress. However, our multivariable analysis showed that diabetes mellitus included in our model was an aggravating factor for ARDS. This may be related to the unique features of COVID-19 infection. COVID-19 potentially uses Angiotensin-Converting-Enzyme 2 (ACE2) as receptor [21], and patients with diabetes mellitus have an increased ACE activity and inflammation [22]. Age and male sex were also included in our model, which suggested that older men with COVID-19 infection were more likely to develop ARDS.
Previous studies have been reporting that LIPS proposed by Gajic et al. could predict the risk of ARDS in the early stages [7,23,24]. According to the LIPS calculation worksheet, higher scores referred to larger risks of ARDS However, compared to the LIPS model, our model had higher AUC, sensitivity, and specificity. Besides, our model included fewer variables, and therefore, it is an easy-to-use predictor index in clinic practice. LIPS was developed to predict ARDS in a large cohort (mainly focused on the major surgery and multiple trauma), and it is not trained for viral infection-induced pneumonia such as COVID-19.
There were some limitations to the study. First, due to the retrospective nature of this study, data were limited to hospital records. Some variables, such as comorbidities, might be underestimated or otherwise inaccurate. Second, we could not predict survival due to the small sample size, which is very important for COVID-19 patients. Future studies should focus on the other clinical outcomes of COVID-19. Thirdly, there was no external validation in our study due to the limited number of centers participating.
Conclusion
We developed and validated the prediction model to provide clinicians and researchers with a simple tool to screen for COVID-19 patients at high risk of ARDS. Early application of this model would be important for deployment in clinical practice. Potential clinical benefits of using this model deserve assessment.
Funding
This work was supported by the National Natural Science Foundation (grants 81772046 and 81971816 to Dr. Peng) and the Special Project for Significant New Drug Research and Development in the Major National Science and Technology Projects of China (2020ZX09201007 to Dr. Peng).
References
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020.
- Phan LT, Nguyen TV, Luong QC, Nguyen TV, Nguyen HT, Le HQ, et al. Importation and Human-to-Human Transmission of a Novel Coronavirus in Vietnam. N Engl J Med. 2020.
- Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, et al. First Case of 2019 Novel Coronavirus in the United States. N Engl J Med. 2020.
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020; 395: 497-506.
- Russell CD, Millar JE, Baillie JK. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet. 2020; 395: 473- 475.
- Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020.
- Gajic O, Dabbagh O, Park PK, Adesanya A, Chang SY, Hou P, et al. Early identification of patients at risk of acute lung injury: evaluation of lung injury prediction score in a multicenter cohort study. Am J Respir Crit Care Med. 2011; 183: 462-470.
- Gattinoni L, Coppola S, Cressoni M, Busana M, Rossi S, Chiumello D. Covid-19 Does Not Lead to a “Typical” Acute Respiratory Distress Syndrome. Am J Respir Crit Care Med. 2020.
- Ferguson ND, Fan E, Camporota L, Antonelli M, Anzueto A, Beale R, et al. The Berlin definition of ARDS: an expanded rationale, justification, and supplementary material. Intensive Care Med. 2012; 38: 1573-1582.
- Sauerbrei W, Royston P, Binder H. Selection of important variables and determination of functional form for continuous predictors in multivariable model building. Stat Med. 2007; 26: 5512-5528.
- Kidd AC, McGettrick M, Tsim S, Halligan DL, Bylesjo M, Blyth KG. Survival prediction in mesothelioma using a scalable Lasso regression model: instructions for use and initial performance using clinical predictors. BMJ Open Respir Res. 2018; 5: e000240.
- Friedman JH, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. Journal of Statistical Software. 2010; 33.
- Trucano TG, Swiler LP, Igusa T, Oberkampf WL, Pilch M. Calibration, validation, and sensitivity analysis: What\”s what. 2006; 91: 1331-1357.
- Bogusz M. Retention of trialkylamines on phenyl methyl silicone capillary columns. J Anal Toxicol. 1988; 12: 239-240.
- Liu J, Liu Y, Xiang P, Pu L, Xiong H, Li C, et al. Neutrophil-to-Lymphocyte Ratio Predicts Severe Illness Patients with 2019 Novel Coronavirus in the Early Stage. medRxiv. 2020: 2020.2002.2010.20021584.
- Liu KD, Matthay MA. Advances in critical care for the nephrologist: acute lung injury/ARDS. Clin J Am Soc Nephrol. 2008; 3: 578-586.
- Mehta RL, Pascual MT, Gruta CG, Zhuang S, Chertow GM. Refining predictive models in critically ill patients with acute renal failure. J Am Soc Nephrol. 2002; 13: 1350-1357.
- Sandhu M, Mukhopadhyay S, Sharma SK. Tropical pulmonary eosinophilia: a comparative evaluation of plain chest radiography and computed tomography. Australas Radiol. 1996; 40: 32-37.
- Thompson BT, Chambers RC, Liu KD. Acute Respiratory Distress Syndrome. N Engl J Med. 2017; 377: 562-572.
- Trillo-Alvarez C, Cartin-Ceba R, Kor DJ, Kojicic M, Kashyap R, Thakur S, et al. Acute lung injury prediction score: derivation and validation in a populationbased sample. Eur Respir J. 2011; 37: 604-609.
- Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS. J Virol. 2020: JVI.00127-00120.
- Srivastava P, Badhwar S, Chandran DS, Jaryal AK, Jyotsna VP, Deepak KK. Imbalance between Angiotensin II - Angiotensin (1-7) system is associated with vascular endothelial dysfunction and inflammation in type 2 diabetes with newly diagnosed hypertension. Diabetes Metab Syndr. 2019; 13: 2061-2068.
- Soto GJ, Kor DJ, Park PK, Hou PC, Kaufman DA, Kim M, et al. Lung Injury Prediction Score in Hospitalized Patients at Risk of Acute Respiratory Distress Syndrome. Crit Care Med. 2016; 44: 2182-2191.
- Kida Y, Ohshimo S, Shime N. Optimal Cutoff Value for Lung Injury Prediction Score and Potential Confounders for Identifying the Risk of Developing Acute Respiratory Distress Syndrome. Crit Care Med. 2017; 45: e624-e625.