
Research Article
Austin J Nanomed Nanotechnol. 2021; 9(1): 1062.
Effect of Gold Nanoparticle Aggregation on the Kinetic Aspect of AuNPs/DNA Interactions
Grueso EM1*, Giraldéz-Pérez RM2 and Prado-Gotor R1
1University of Seville, Department of Physical Chemistry, Spain
2University of Cordoba, Faculty of Science, Department of Cellular Biology, Physiology and Immunology, Spain
*Corresponding author: Grueso EM, Department of Physical Chemistry, University of Seville, Spain; Email: elia@us.es
Received: February 05, 2021; Accepted: March 03, 2021; Published: March 10, 2021
Abstract
Since successful therapy for curing cancer and others genetic diseases requires the transport of DNA into the cell by delivery vehicles, the understanding of the factors that control the complexation and condensation of the DNA is a key problem. During the last decade, researchers have developed some uses of nanoparticles-DNA systems, the majority of these studies dealing with NPs, which are covalently bound to the DNA. However, the kinetic aspect of AuNPs/DNA system by non-covalent interactions is less explored. Moreover, the role of high salt concentrations in these studies is of great interest due to the majority of nanoparticles have a great tendency to aggregate upon exposure to biological medium, significantly alter the uptake extent, rate, and mechanism of AuNPs/DNA interaction. As a contribution to this field, we have studied kinetics aspects of the binding of small tiopronin gold nanoparticles, AuNPs, to double stranded DNA in at high salt concentration by using the stopped-flow technique. The kinetic curves are biexponential and reveal the presence of two kinetic steps. Moreover, AFM studies reveal AuNPs aggregation in the presence of high salt content, while the same particle are well-dispersed in water. A two-step series mechanism reaction scheme was proposed. According to the reaction scheme, the formation of an intermediate complex formed by aggregated gold nanoparticles and DNA precedes the rate-determining step of the reaction.
Keywords: DNA; Gold nanoparticles; Kinetic; AuNPs-aggregation
Introduction
The understanding and dealing of nanoparticle/DNA interactions has become an important emerging area of research due to the high number of diagnostic and therapeutic applications derived from these systems [1-4]. In particular, the comprehensive study of DNA-AuNPs affinity interactions have contributed to the promising challenge of the use of these systems to treat diseases, specifically, hereditary diseases by the insertion of genes into the human cells is so called gene therapy. Importantly, the gene transfection strategies need, as a pre-requisite, the effective complexation and the collapse of extended DNA chains into compact DNA structures [5]. Of particular note, non-covalent interactions between nanoparticles and DNA have been recognized to control physicochemical aspects of the interaction. In this sense for each nanoparticle-DNA systems a global effect is exerted in conjunction by both the metal cluster core and the capping agents that contribute to stabilize the nanosystem [6]. Regarding the metal cluster core, especially electrostatic interactions [7,8], hydrophobic forces [9], and the specific bonding between the chemical groups of DNA bases and the metal center control these interactions [10].
In relation to this, understanding the interactions of aggregates or isolated nanoparticles with DNA is determinant to their use in vivo delivery of drugs and efficacious nanomedicine design. In fact, the exploration of new nanoparticle systems that are capable of changing the aggregation state within the physiological environment constitutes an emerging concept [11]. In a previous paper, the effect of etanol on the kinetic of aggregation of AuNPs/DNA was explored, revealing not only a change in the binding mode induced by the solvent but also on the mechanism of its interaction [12]. However, the effect of high salt concentration on AuNPs/DNA mechanism is unexplored. Since nanoparticles may aggregate upon exposure to biological medium, due to the presence of aggregation-inducing molecules/species (such as salt), exploring effect of high salt concentration on the kinetic and thermodynamic of AuNPs/DNA interactions is a topic of great interest. In fact, the presence of high salt concentration may significantly alter the uptake extent, rate and mechanism of AuNPs/DNA interaction [13]. In a previous work, the interaction of small tiopronin gold nanoparticles, Au@tiopronin, with DNA biopolymer was evaluated at very low [NaCl] (0.001-0.015 M) [14]. The result demonstrated that the kinetic results are compatible with a three-step series mechanism reaction scheme, in which the groove binding interactions of DNA and gold nanoparticles was governed by solvation and viscosity factors [14]. In this study we have investigated the effect of high salt concentrations on the kinetic of Au@tiopronin with DNA. The simplest mechanism consistent with the kinetic results involves a more simple reaction scheme with two-step reactions. The first step corresponds to a very fast step that is related to a diffusion controlled formation of an external precursor complex between aggregated gold nanoparticles and DNA. The second step involves the formation of an external complex, as a result of the binding affinity between hydrophilic groups of the aggregated tiopronin nanoparticles and the DNA grooves. As a summary, this study reveals that kinetic of AuNPs/DNA interaction is controlled by AuNPs aggregation state, revealing the importance to take great care in control of the solvent media for diagnostic and therapeutic applications derived from these systems.
Materials and Methods
All chemicals were of Anal. R. grade and were used without further purification. Hydrogen tetrachloroaureate (III) trihydrate, 3-Aminopropyltriethoxilane (APTES), NaCl and BaClO4 were purchased from Sigma-Aldrich; N-(2-mercaptopropionyl)glycine from Fluky; NaBH4 from Lancaster. Calf thymus DNA was purchased from Pharmacia and used without further purification, because preliminary experiments showed that purification does not produce any changes in the experimental results. The absorbance ratio of DNA stock solutions at 260nm and 280nm was monitored and found to be between 1.8 and 1.9 (A260/A280 = 1.87), indicating no protein contamination [15]. An agarose gel electrophoresis test using ethidium bromide indicated that the average number of base pairs per DNA molecule is above 10,000 bp [16]. Polynucleotide concentrations were determined spectrophotometrically from the molar absorptivity (6600M-1cm-1 at 258nm in order to have the DNA concentration in phosphate units) [17]. Solutions were prepared with de-ionized water, its conductivity being less than 10-6Sm-1. Tiopronin gold nanoparticles, Au@tiopronin were synthesized by using Templeton et al.’s procedure [18]. Gold nanoparticles were characterized by visible absorption spectra, TEM and microanalysis (11.8% C; 1.86% H; 2.89% N; 5.80% S; Au 70.88%, C410H656O246N82S82Au197). A value of (1.6 ± 0.2) nm was obtained for the diameter of the gold nanoparticle in the absence of any added salt (Figure 1). According to these data, the relationship between the number of Au atoms and tiopronin ligands was 197/82.
Kinetics
The kinetic experiments were performed at 298.0K by using a Biologic SF 300 stopped-flow instrument and monitoring the course of the reaction in the CD detection mode. This detection mode was employed because the signal-to-noise ratio was found to be more favorable compared to the absorbance mode. The acquired signal was recorded on a PC and then analyzed by using the Jandell AISN software program. The Au@tiopronin concentration was 1.0×10-6M in all experiments, and the DNA concentration was varied. All the kinetic experiments were performed under pseudo first-order conditions ([DNA] >10 [Au@tiopronin]). Each experiment was repeated at least 10 times, and the relevant kinetic traces were accumulated in order to reduce the signal-to-noise ratio. The spread of time constants was found to be within 10%.
TEM measurements
For TEM visualization, a single drop (10mL) of an AuNPs aqueous solution was placed on a carbon film coated copper grid, which was then left to air dry for 2 hours at room temperature. TEM analysis was carried out using a Philips CM 200 electron microscope working at 200kV, and the resulting images were analyzed by using Image J free software.
Circular Dichroism (CD) spectra
Electronic CD spectra were recorded using a BioLogic Mos-450 spectropolarimeter. A standard quartz cell of 10mm path length was used. The spectra were expressed in terms of molar ellipticity. Scans were taken from 220nm to 310nm for the intrinsic region. For each spectrum, 5-10 runs were averaged at a constant temperature of 25.0ºC, with a 5min equilibration before each scan. All the spectra were recorded at a fixed concentration of double-stranded DNA, CDNA = 1.0×10-4M, and CNaCl = 0.50M.
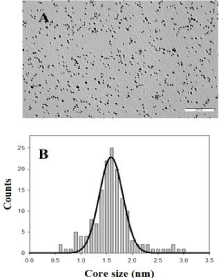
Figure 1: TEM image of Au@tiopronin gold nanoparticles in water solution
and the corresponding histogram of size distribution.
AFM measurements
Images were obtained in air and in tapping mode using a Molecular Imaging PicoPlus 2500 AFM (Agilent Technologies). Silicon cantilevers (Model Pointprobe, Nanoworld) with a resonance frequency of around 240kHz and a nominal force constant of 42Nm-1 were used. All AFM images were recorded with scan speeds of about 0.5Hz, and data were collected in a 256×256 pixel format. For AFM imaging of AuNPs in water and in the presence of high salt, a 100µL droplet of the sample was deposited on a freshly cleaved mica surface, which had been previously modified with 0.1% (v/v) APTES solution, and incubated for 40min. The surface was then washed with ultrapure water and finally air-dried. All images presented here are the original data, except that a flatten command was used to remove the background slope [19].
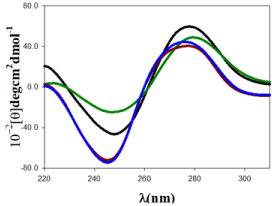
Figure 2: CD spectra of Au@tiopronin/DNA systems in different experimental conditions, CDNA = 100µM in all the spectra. (A) CAu@tiopronin = 0.0µM and CNaCl = 0.0M, black spectrum; (B) CAu@tiopronin = 5.0µM and CNaCl = 0.0M, green spectrum; (C) CAu@tiopronin = 0.0µM and CNaCl = 0.5M, blue spectrum; (D) CAu@tiopronin = 5.0µM and CNaCl = 0.5M, red spectrum.
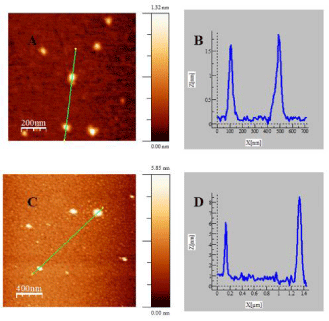
Figure 3: AFM topographic images of Au@tiopronin gold nanoparticles in
water (A) and NaCl: 0.5M (C). Figures B and D correspond to cross sectional
analysis of the heights along the selected lines for images (A) and (C),
respectively.
Result and Discussion
Small Au@tiopronin gold nanoparticles induce a conformational change in double stranded DNA upon binding which can be followed by the circular dichroism technique (Figure 2). Double stranded DNA shows a CD spectrum characteristic of the right-handed B-form in the far UV region (220-320 nm). Structural alterations of the biomolecule caused by its interaction with gold nanoparticles as ligand are reflected in changes in this intrinsic CD spectrum. Figure 2 (black colour) shows a CD spectrum with the characteristic features of ds-DNA in B-form, having a positive peak at 278nm and a negative peak at 247nm. These bands arises by stacking interactions and the helical superstructure of the polynucleotide that provide an asymmetric environment for the bases [20]. Upon the addition of 5.0µM of Au@tiopronin gold nanoparticles to the DNA in water solution, the molar ellipticity decreases at approximately 278nm, and increases at approximately 247nm. These changes are coupled with a shift in the maximum wavelength of the positive band, indicating partial denaturation and the double helix unwinding (Figure 2, green colour) [14]. However, the effect of adding high salt concentration in DNA structure, both in the presence and in the absence of gold nanoparticles, is thoroughly different (Figure 2, red and blue colour). Namely, when CNaCl = 0.5M was added to the solution both bands are displaced to a lower wavelength and a decrease in the intensity of positive band together with an increase in the intensity of the negative CD band is registered. Note that this change in CD spectra is compatible with a conformational change from B to C-form of DNA, which is in progress due to changes in solvent conditions. In fact, the conformational change from B to C-DNA form causes a decrease in a specific CD band as well as the progressive dehydration of DNA as salt concentration increases; thus, the shrinkage of the double helix yields more condensed or packed DNA structures [21]. It is important to note that increasing in the size and depth of the major groove, which accompanies this transition, may be important for the mode of binding that tiopronin gold nanoparticles experience in its interaction with DNA [12]. In fact, Figure 2 shows that DNA undergoes a less pronounced change in CD spectra, when CAu@tiopronin = 5µM was added to the DNA solution, in the presence of high salt concentration. Note that this behavior could be compatible with an external binding of gold nanoparticles to the biomolecule.
To explore the effect of adding high salt on Au@tiopronin aggregation state, AFM experiments were carried out in the absence and in the presence of NaCl (Figure 3). AFM measurements in water reveal the presence of isolated Au@tiopronin (Figure 3A-3B) with a mean diameter of 1.6 ± 0.1 nm in z-direction, in good agreement with TEM measurements (Figure 1). However, AuNPs morphology is modified at CNaCl = 0.5M (Figure 3C-3D), visualizing different z-aggregates with a mean diameter of 6.5 ± 1.6 nm, revealing that an average of 4 nanoparticles constitutes the aggregate in salt. This finding is relevant for possible biomedical applications of Au@tiopronin/DNA system. Thus, according to Albanese et al. the uptake patterns in different cell lines is different between single and aggregated nanoparticles, revealing the importance of AuNPs-aggregation in mediating targeting and intracellular trafficking [22].
As regards the kinetic study of Au@tiopronin binding to DNA in the presence of high salt concentration, the kinetic curves were appropriately fitted by two exponentials, whose relaxation times, τ1 and τ2, differ by about 1 order of magnitude. An example of a typical kinetic experiment appears in Figure 4. The kinetic behavior of the system can be rationalized on the basis of the two-step series model depicted in Scheme I:
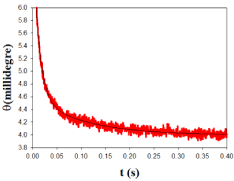
Figure 4: Plot of ellipticity vs time, t, in a typical kinetic experiment. The dots
represent experimental values, and the curve is the best fit of the experimental
data to a biexponential equation.
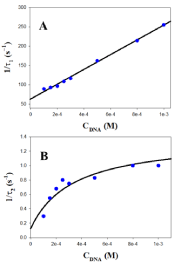
Figure 5: Reciprocal fast (1/τ1, A) and slow (1/τ2, B) relaxation times dependence on DNA concentration at 298.0K in the presence of [NaCl] = 0.5M.
Salt type
K ( M-1)
K1 (M-1)
K2
k1 (M-1s-1)
k-1 (s-1)
k2 (s-1)
k-2 (s-1)
[NaCl] = 0.5M
34.7×103
3.0×103
10.5
1.91×105
63
1.3
0.12
[BaClO4] = 0.5M
4.0×103
7.0×102
4.7
2.05×104
29
4.4
0.93
Table 1: Equilibrium and Rate Constants for the Interaction of DNA with Au@tiopronin in the presence of high salt concentrations.
where (DNA/(AuNPs)AGG)I and (DNA/(AuNPs)AGG)I represent two different DNA-AuNPs complexes and (AuNPs)AGG corresponds to the Au@tiopronin aggregates formed at high salt content. A comment about the change in the mechanism of interaction of Au@tiopronin with DNA respect to the same system studied in water and low NaCl content (from 0 to 0.015 M) seems to be pertinent. Thus, the three-step mechanism observed at low salt evolves to a two-step series mechanism when salt concentration increases [14]. In this study, the dependence of the reciprocal of the fast relaxation time, 1/τ1, on the DNA concentration is linear, according to eq. 1 [23].
Therefore, individual k1 and k-1 kinetic constants can be obtained from the plot of 1/τ1 versus the DNA concentration. Examples of this kind of plot are shown in Figure 5A and 6A in the presence of 0.5M of NaCl and BaClO4, respectively. Subsequently, once k1 and k-1 have been obtained from eq. 1, the value of the individual equilibrium binding constant of the first step, K1, is calculated as k1/k-1. Moreover, Figure 5B and 6B show that the dependence of the reciprocal slow relaxation time, 1/τ2, on the DNA concentrations is curve and tends to a plateau in agreement with eq. 2 [23].
The value of K1 obtained from equation 1 as k1/k-1 is introduced into eq. 2 as a known parameter, while k2 and k-2 are evaluated by data fit to eq. 2 (Figure 5B and 6B). Subsequently, the value of K2 is calculated as k2/k-2. Once individual kinetic and thermodynamic parameters are calculated, the value of the global equilibrium binding constant is obtained as follow from K = K1×(1+K2); the reaction parameters obtained are collected in (Table 1).
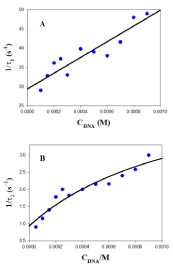
Figure 6: Reciprocal fast (1/τ1, A) and slow (1/τ2, B) relaxation times dependence on DNA concentration at 298.0K in the presence of [BaClO4] = 0.5M.
Concerning the stability of the intermediate complex (DNA/(AuNPs)AGG)I, it can be hypothesized that the binding could be mediated fundamentally by non-electrostatic interactions in accordance with other uncharged ligand/DNA systems [16]. This is due to in the present system and reaction condition, Au@tiopronin nanoparticles are slightly anionic and hydrophilic [14]; thus, the electrostatic interactions could be neglected and the stability of the external complex could be mediated by hydrogen bonding between the hydrophilic groups of the tiopronin chain and the DNA bases. Moreover, Table 1 shows that k1 and k-1 decreases as increases the ionic strength in the media from NaCl 0.5M (I = 0.5M) to BaClO4 (I = 1.25M), which is in accordance with the idea that a progressive dehydration of the DNA occur in the transition from B to C-like form as ionic strength increases. Therefore, this fact could hinder the hydrophilic interactions that are needed to stabilize the (DNA/(AuNPs)AGG)I complex as ionic strength increases.
On the other hand, concerning the variations of k2 and k-2 with the ionic strength, it could reasonably suppose that a reorganization of the (DNA/(AuNPs)AGG)I complex to a more compacted form, (DNA/(AuNPs)AGG)II, governs the rate-determining step of the reaction and its reverse process. Note that the variation tendency of k2 and k2 with ionic strength is the opposite than that observed for k1 and k-1 individual constants. This finding highlights that the formation of the final packed DNA/AuNPs structures is favored at high ionic strength due to the starting DNA conformation is closer to the final condensed stage.
Finally, it is worthy of note the values obtained for the global equilibrium binding constant, K = 3.47×104 and 4.0×103M-1, in the presence of 0.5M of NaCl and 0.5M BaClO4, respectively, are greater taking into account the high salt condition. In fact, comparing the K values for DNA/AuNPs system collected in (Table 1) with those obtained for 1-PyCHO/DNA system, in which the dye is also of neutral character as in the case of Au@tiopronin, (K = 1.0×104 at 0.5M of NaClO4 and 3.0×103 at 0.5M of BaClO4), it is obvious that the formation of DNA/AuNPs complexes is optimal at the selected high ionic strength [24]. Note that binding constants values in the 104-106 range, as that found here at high NaCl content, are reported as optimal for the complexation of the ligands and the consequent release once reached the target site [25]. Specifically, in the context of cellular applications, it was demonstrated that a high value of the DNA/AuNPs binding constant would leads to better intracellular binding of the target molecule, thereby increasing the effectiveness of antisense gene regulation [26]. The favorable interaction between Au@tiopronin and ds-DNA explored at high salt can therefore support the bioavailability of the system in physiological environment, for possible medical applications.
Conclusion
Details on the kinetic and thermodynamic of the binding of small tiopronin gold nanoparticles to double stranded DNA in the presence of high salt have been here provided through the use of stopped-flow and CD spectroscopic techniques. Moreover, the aggregation state of nanoparticles was analyzed by using the ultrasensitive AFM technique. The proposed mechanism consistent with analysis of the data provided involves a two-step series reaction scheme. In the first step, aggregated gold nanoparticles forms an intermediate external complex with DNA mediated by non-electrostatic interaction. Then, in the second step, a more DNA/AuNPs packed structure is formed, which is favored kinetically at high ionic strength. On the whole, we can say that this study contributes to obtaining a deeper comprehension of the uptake patterns in cell in which the DNA/AuNPs are involved. The change of binding mode and mechanism that DNA/AuNPs system undergo at high salt content reveals the importance of study AuNPs aggregation in mediating targeting and intracellular trafficking of DNA/AuNPs systems.
Acknowledgments
This work was financed by the University of Seville, Spain V PP USO SSGG (2019/00000570) and VI PP USO SSGG (2020/00001068). The authors are grateful to the Microscopy and Functional Characterization Service of the Research, Technology and Innovation Center of the University of Seville (CITIUS).
References
- Prow TW, Grice JE, Lin LL, Faye R, Butler M, Becker W, et al. Nanoparticles and Microparticles for Skin Drug Delivery. Adv Drug Delivery Rev. 2011; 63: 470-491.
- Ghosh PS, Han G, De M, Kim CK, Rotello VM. Gold Nanoparticles in Delivery Applications. Adv Drug Delivery Rev. 2008; 60: 1307-1315.
- Ma W, Chen M, Kaushad S, McElroy M, Zhang Y, Ozkan C, et al. PLGA Nanoparticle-Mediated Delivery of Tumor Antigenic Peptides Elicits Effective Immune Responses. Int J Nanomed. 2012; 7: 1475-1487.
- Zhou T, Llizo A, Wang C, Xu G, Yang Y. Nanostructure-Induced DNA Condensation. Nanoscale. 2013; 5: 8288-8306.
- Davis ME. Non-Viral Gene Delivery Systems. Curr Opin Biotechnol. 2002; 13: 128-131.
- Grueso EM, Perez-Tejeda P, Prado-Gotor R and Cerrillos C. DNA Strand Elongation Induced by Small Gold Nanoparticles at High Ethanol Content. J. Phys. Chem. C. 2014; 118: 4416-4428.
- Zhang X, Servos MR, Liu J. The Surface Science of DNA Adsorption onto Citrate-Capped Gold Nanoparticles. Langmuir. 2012; 28: 3896-3902.
- Herne TM, Tarlov MJ. Characterization of DNA Probes Immobilized on Gold Surfaces. J Am Chem Soc. 1997; 119: 8916-8920.
- Nelson EM, Rothberg LJ. Kinetics and Mechanism of Single-Stranded DNA Adsorption onto Citrate-Stabilized Gold Nanoparticles in Colloidal Solution. Langmuir. 2011; 27: 1770-1777.
- Dremers ML, Ostblom M, Zhang H, Jang NH, Liedberg B and Mirkin CA. Thermal Desorption Behavior and Binding Properties of DNA Bases and Nucleosides on Gold. J Am Chem Soc. 2002; 124: 11248-11249.
- Zagorovsky K, Chou LYT, Chan WCM. Controlling DNA-nanoparticle serum interactions. Proc Natl Acad Sci. USA. 2016; 113: 13600-13605.
- Grueso EM, Pérez-Tejeda P, Giraldéz-Pérez RM, Prado-Gotor R, Muriel-Delgado F. Ethanol effect on gold nanoparticle aggregation state and its implication in the interaction mechanism with DNA. J Colloid Interface Sci. 2018; 529: 65-76.
- Alkilany AM, Mahmoud NN, Hashemi F, Hajipour MJ, Farvadi F and Mahmoudi M. Misinterpretation in Nanotoxicology: A Personal Perspective. Chem Res Toxicol. 2016; 29: 943-948.
- Prado-Gotor R, Grueso EM. A kinetic study of the interaction of DNA with gold nanoparticles: mechanistic aspects of the interaction. Phys Chem Chem Phys. 2011; 13: 1479-1489.
- Sambrook J, Fritsch EFF and Maniatis T. Molecular Cloning: A Laboratory Manual. 4th edition. New York: Cold Spring Harbor Laboratory Press. 1989.
- Secco F, Venturini M, Biver T, Sanchez F, Prado-Gotor R, Grueso EM. Solvent Effects on the Kinetics of the Interaction of 1-Pyrenecarboxaldehyde with Calf Thymus DNA. J Phys Chem B. 2010; 114: 4686-4691.
- Felsendeld G, Hirschman SZ. A neighbor-interaction analysis of the hypochromism and spectra of DNA. J Mol Biol. 1965; 13: 407-427.
- Templeton AC, Chen S, Gross SM, Murray RW. Water-Soluble, Isolable Gold Clusters Protected by Tiopronin and Coenzyme A Monolayers. Langmuir. 1999; 15: 66-76.
- Horcas I, Fernández R, Gómez-Rodríguez JM, Colchero J, Gómez-Herrero J, Baro AM. WSXM: a software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instrum. 2007; 78: 013705.
- Neidle S. Nucleic Acid Structure and Recognition. Oxford University Press. New York. 2002.
- Ranjbar B, Gill P. Circular Dichroism Techniques: Biomolecular and Nanostructural Analyses- A Review. Chem Biol Drug Des. 2009; 74: 101-120.
- Albanese A and Chan WCW. Effect of Gold Nanoparticle Aggregation on Cell Uptake and Toxicity. ACS Nano. 2011; 5: 5478-5489.
- Bernasconi CF. Relaxation Kinetics. New York: Academic Press. 1976.
- Grueso E, Sanchez F, Martin VI, García-Fernández E, Prado-Gotor R. Quantification of salts and cosolvents–DNA interactions in terms of free energies: A study using the pyren-1-carboxyaldehyde as fluorescent probe. Chemical Physics. 2008; 352: 306-310.
- Topala T, Bodoki A, Oprean L, Oprean R. Bovine serum albumin interactions with metal complexes. Clujul Med. 2014; 87: 5.
- Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006; 312: 1027-1030.