
Research Article
Austin J Microbiol. 2024; 9(1): 1045.
Development of Covid-19 Agglutination Test Using Antibody-Sensitized Latex Beads
Noor-US-Saba¹; Khalida Iqbal²*; Rashida Bano²; Faiza Jamil²; Naveed Ali²; Munawwar Shah²; M Owais Qadri²; Fayyaz Ahmed²; Affan Tahir²; Saifullah Khan²
¹Department of Microbiology, University of Karachi, Karachi, Pakistan
²Centre of Excellence in Science and Applied Technologies, Islamabad, Pakistan
*Corresponding author: Khalida Iqbal Centre of Excellence in Science and Applied Technologies, Islamabad, Pakistan. Email: khalidaiqball@gmail.com
Received: December 15, 2023 Accepted: January 27, 2024 Published: February 03, 2024
Abstract
Objective: To develop an effective and time-saving method using the latex agglutination technique for the detection of SARS-CoV-2 in nasopharyngeal swabs.
Method: Rabbits were immunized with inactivated COVID-19 vaccines to generate anti-SARS-CoV-2 antibodies. These antibodies were coated on 0.8μm polystyrene latex beads by adsorption method. Then, slide agglutination tests were performed using COVID-19 PCR samples which were then further verified with light microscopy of the agglutinants. Additionally, Digital microscopy and electron microscopy were used for validation of the results. Furthermore, convolutional Neural Network (CNN), a machine learning approach was utilized as well to classify the images as positive and negative for the assessment of the potential of the agglutination test as a rapid test for mass screening.
Results: Slide agglutination produced minute aggregates within 4-5 minutes that were further visualized under light and digital microscope which showed a white patchy pattern and shiny aggregates respectively. Electron microscopy was done to further confirm the findings of the assay which revealed a humpy formation or protrusions at the places where antigen and antibody binding took place. Additionally, the CNN model was constructed to predict light microscopy images as positive or negative thus defining the potential of agglutination assay as a point-of-care testing assay.
Conclusion: We have developed a simple testing method based primarily on a microscopic approach for the diagnosis of COVID-19. This technique can be used as a point-of-care testing method as well as for screening purposes in a large population where expensive machinery and time-consuming testing techniques cannot be employed.
Keywords: SARS-CoV-2 detection; Latex agglutination; Slide agglutination; Light microscopy; Digital microscopy; Electron microscopy; Agglutination assay; Antibody-conjugation
Introduction
COVID-19 is a disease caused by a novel coronavirus that belongs to the genus of betacoronaviruses. Its first case was reported in the Chinese capital city of Wuhan [1]. It emerged as a life-threatening disease which was later classified as a pandemic by WHO in 2020 [2]. Clinically it is manifested as fever being one of the first signs, body aches, loss of taste and smell with pneumonia and acute respiratory distress syndrome in severe case [3]. It is a highly contagious disease which spreads through respiratory droplets and aerosols which can remain in the air for an extended period of time especially in places with inadequate ventilation [4]. So it was a challenge for the health care system worldwide to combat this deadly disease as there were no diagnostic and treatment approaches available at that time. As the pandemic became more severe, need for the proper diagnostic approaches increased for timely management of the disease. RT-PCR was the most commonly used and a gold standard method to diagnose active infection, but due to the time constraint and high cost related to this test, demand for rapid diagnostic devices increased for prompt diagnosis and mass screening purposes [5,6]. Among the rapid detection approaches, serological-based techniques for detecting antibodies against COVID-19 disease were employed, including lateral flow assays and ELISA [7]. Latex Agglutination-based assays are also one of the commonest and cheapest methods to attain fast and accurate results [8]. This technique is based on coupling antibodies or antigens with the polystyrene latex beads, which generate clumps when interacting with their corresponding antigens. Many commercially available diagnostic tests are based on this technique due to its cost-effectiveness and convenience [9].
In the current study, we also aimed to develop a fast and accurate method based on latex agglutination technique to detect SARS-CoV-2 in nasal swabs. The advantage of this assay is its ability to diagnose current infection by detecting viral antigens rather than past infection which is based on antibody detection. So, this assay can be used as an inter-dependent method for the detection of COVID-19 when dealing with a large number of samples or in patients’ screening.
Methodology
Ethical Approval
Prior to the commencement of the present study, the study protocol was thoroughly reviewed and approved by the Institutional Bioethics Committee to ensure ethical soundness and compliance with local, national, and international guidelines for animal research.
Animal Immunization
Two groups of rabbits were immunized with four doses of commercial inactivated COVID-19 vaccines. Doses were given every week for one month. Subsequently, blood was collected thrice after the administration of the last dose with an interval of one week between each bleed. Then, the assessment of immune response was done by employing indirect serum ELISA and viral neutralization assay (Data not shown).
Purification of Covid-19 Polyclonal Antibodies
Sera with the potent immune response were selected for the purification of antibodies using the Protein A column to purify the polyclonal antibodies. Afterward, SDS-PAGE of purified antibody fractions was performed to confirm and analyze purified IgG followed by dialysis against PBS for 5 days at4C. After dialysis, all the antibodies were concentrated using a concentrator of 30kDa. To characterize and confirm the purity of antibodies in concentrate, the SDS-PAGE was run again in both reducing and non-reducing conditions. The concentration of total proteins in the concentrated fractions was estimated using the Bradford method and antibody characterization was done by western blot analysis (Data not shown).
Coating of Antibodies on Latex Beads
Antibodies were coated on deep-blue dyed polystyrene latex beads of size 0.8μm by employing the adsorption technique. For this, the microsphere-to-protein ratio to achieve surface saturation was calculated by following Bang’s laboratories’ adsorption to microsphere protocol [10]. In the first step, latex beads suspended in borate buffer were added to the calculated amount of SARS-COV-2 polyclonal antibodies followed by overnight incubation at 4°C. Then, blocking was done with 0.5% BSA and PBS along with for 2 hours after washing the sensitized beads twice with 0.05% BSA with coat buffer by centrifugation at 8000 rpm for 5 minutes. Lastly, antibody-coated beads were stored in PBS and 1% glycerol at 4 degrees till further use.
Slide Agglutination Assay with Live Samples
SARS-COV-2 positive patients’ nasopharyngeal swab samples detected by RT-PCR were assessed to visualize the agglutination with antibody-sensitized latex beads. For this purpose, conjugated beads were added to the swab sample i.e. 10μl of SARS-CoV-2 positive sample added to a clean glass slide followed by the addition of 10μl of latex beads and mixing using a sterile disposable wire loop and let the reaction proceed for 3-5 minutes. A blank reaction without the addition of a sample was also performed on the same slide to detect false positive results. All the safety measures were adapted to carry out the reactions under BSC-II.
Light Microscopy of Agglutination
To observe the viral agglutination under a microscope for better visualization and understanding of the formed agglutinants, light microscopy under a 100X lens was performed after allowing the slides to be air-dried. It was seen that the air-dried slides showed a complex pattern of networking in patients’ samples, so further analysis was done under the microscope.
Digital Microscopy
As light microscopy requires expertise and experienced personnel to handle the microscope due to its sophistication and expensiveness, we switched to a digital microscope which is cheap and easy to use to further visualize and confirm our findings and to make this method more approachable. For this, Air-dried slides were seen through a digital microscope by placing them over the lens. Visualization was done using a computer screen which has the software already installed for the microscope.
Electron Microscopy
In order to validate and further confirm our results from digital and light microscopes, electron microscopy was done to visualize the aggregates. Samples were prepared on a small piece of glass slide in a 1:1 ratio with conjugated beads. The slide sections were allowed to air-dry inside the biosafety cabinet and then further processed for the microscopic analysis.
Construction of Neural Network
The dataset generated from the light microscopy was used to train the Convolutional Neural Network (CNN). CNN is widely used in machine vision and medical image analysis. The dataset comprises of 73 instances which include 57 images of agglutination reaction between virus and antibodies and 16 images of negative agglutination reaction and split into train, test and validation. Since the data size is limited, new instances have been generated using Open CV and Keras image preprocessing module. Dataset augmentation is usually carried out to enhance the performance of CNN and decrease the chances of overfitting. Therefore, the images present in the train dataset were augmented to significant numbers by incorporating different steps which include rotation of images to 40 degrees, brightness adjustment, zooming and flipping of images using Keras Image Data Generator library. Furthermore, calculated Weights have been assigned to positive and negative instances to balance the data [11].
A sequential model of CNN network was constructed by incorporating 3 convolutional layers, 2 max-pooling layers, and 2 fully connected layers. The input size of the image was set to 128x128 pixels and ReLU was used as an activation function. Finally, a sigmoid layer containing 2 neurons was applied to classify the images into two different classes i.e. positive and negative. Meanwhile, a sequential CNN model based on transfer learning was also implied using pre-trained models i.e. VGG-16, ResNet, and Xception on imageNet dataset as it is mentioned in literature that transfer learning with ImageNet dataset which is a non-medical images dataset works best with medical dataset as well [12].
Validation of the Assay
The above established immunodiagnostic assay was validated using 18 PCR negative and 20 PCR positive samples to see any discrepancy in the results. Samples were obtained from commercial diagnostic laboratories with their reports to remove any source of doubt so that validation can be justified. Positive Samples with the Cycle Threshold (Ct) value ranging from 19-34 were chosen for the validation purpose, then sensitivity and specificity was calculated using mathematical formulae given below:
Sensitivity = True positive/ True positive + false negative
Specificity = True negative/ True negative + false positive
Results
Sds-Page Analysis of Purified Antibodies
To determine the purity of IgG antibodies after affinity column purification, SDS PAGE was performed with the results represented in Figure A showing a band size between 180-245kDa in non-reducing conditions corresponding to the presence of intact antibodies without the breakage of di-sulphide bonds while in reducing conditions bands at 48 and 25kDa are observed which indicates the presence of IgG heavy and light chains respectively.
Performance of Latex Agglutination Assay
When sensitized beads were added to the sample and mixed followed by continuous rotation of the slide, agglutination was observed after 6-10 minutes approximately in the positive samples while t it was seen that there was clear background with the absence of any sort of agglutination in the blank as well as in PCR negative samples.
Microscopic Analysis of Agglutinants
As the size of the clumps was very small, so microscopic analysis was done for better visualization and confirmation of the results. Air-dried slides were observed under the 100x lens of light microscope which gave a white patchy appearance under a microscope corresponding to the agglutination while there was no such color change or white-patches formation in blank and negative samples.
Digital Microscopy
After light microscopy, to further make this detection technique more user-friendly and testing its capability to be modified as an easy-to-handle and rapid diagnostic test, a digital microscope was used which has a magnification of 1600x.
Blank and negative samples under digital microscope: A blank sample containing a coating buffer and sensitized beads was visualized under the digital microscope which was seen as a dark blue area only (Figure 7). On the other hand, negative samples were also seen as a deep blue area corresponding to the presence of blue latex beads only and indicating the absence of viral antigens (Figure 8).
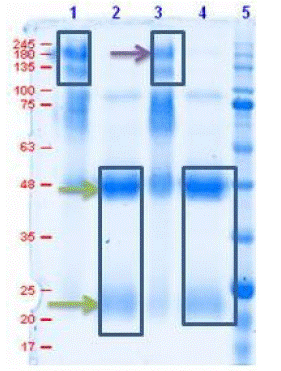
Figure 1: SDS-PAGE of purified antibodies in reducing and non-reducing condition.
Lane 1 & 3: Antibodies under non-reducing cond.
Lane 2 & 4: Antibodies under reducing cond.
Lane 5: protein marker (MOLEQULE-ON)

Figure 2: Reaction of conjugated beads with PCR-negative samples and blank. a, c and e): Blanks- Borate buffer with sensitized beads. b, d and f): Negative samples with sensitized beads.
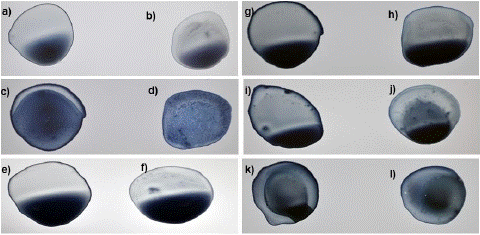
Figure 3: Reaction of conjugated beads with blanks and PCR-positive samples of different Ct values. a,c,e,g,I and k): Blanks- Borafe buffer with sensitized beads. No granules or network formation. b) Small sized clumps or aggregates in sample with Ct-value 19 d) formation of granular network in sample with Ct-value 22. f) formation of very small aggregates in sample with Ct-value 24. h) very minute sized granular formation in sample with Ct-value 26. j) few aggregates at discrete places in sample with Ct-value 28. l) undefinable or insignificant aggregation in sample with Ct-value 30.
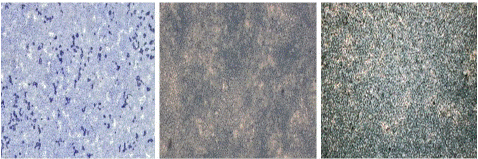
Figure 4: “Blank” samples seen under light microscope at 100x magnification.

Figure 5: “Negative” samples seen under light microscope at 100x magnification.
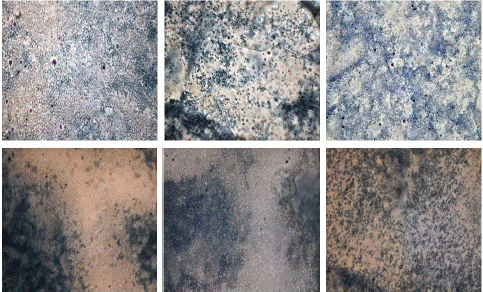
Figure 6: “Positive” samples with different Ct-values seen under light microscope at 100x magnification appearing as white patches formation.

Figure 7: “Blank” seen under digital microscope.
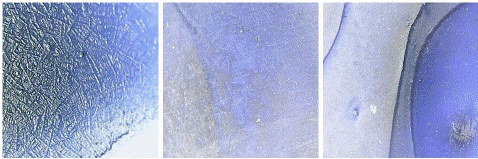
Figure 8: “Negative” seen under digital microscope.
Positive samples under digital microscope: Aggregation or clump formation in positive samples were seen like bright and shiny crystals spread all over the surface. There was a significant different between the boundaries of positive and negative samples as the latter had plain or empty boundaries while the former had those clumpy areas at that place (Figure 9).

Figure 9: “Positive samples” seen under digital microscope.
Electron Microscopy
To validate the findings of light microscopy and digital microscope, electron microscopy was performed for negative and positive samples. Under an electron microscope, negative samples were seen as antibody-conjugated beads only, there was no indication of clumping or aggregation while protrusions of aggregations were seen in positive samples indicating the clump formation by antigen and antibody interactions (Figure10).
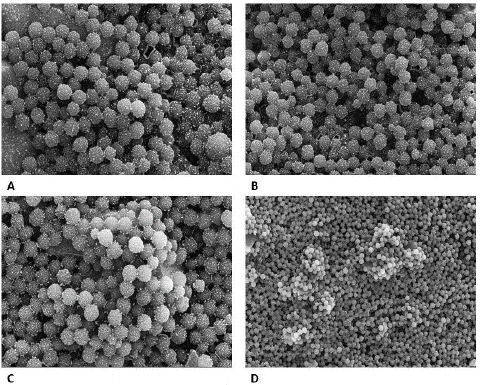
Figure 10: “Negative” (A & B) and “Positive samples” (C & D) seen under Electron microscope. A: scale- 2μm, Mag-30,000x, B: Scale - 3μm, Mag- 25000x. C: Scale- 2μm, Mag: 30,000x. D: Scale- 5μm, Mag: 10,000x.
Application of Artificial Intelligence (AI) on Image Classification
The CNN sequential models having different convolutional layers and number of nodes were generated to classify the light microscopy images. The images were augmented using OpenCV and Keras image preprocessing module of keras deep learning library. A total of 5 models trained with high accuracy were selected. The models were implied on validation dataset comprises of 20 positive and 16 negative instances which also include blank samples i.e., antibody coated beads with media as well. Model E that has been trained with augmented images using keras library of image augmentation works well both in terms of specificity and sensitivity and has been selected for future implementation. While models i.e., Model B, Model C, Model D trained on pretrained models like Xception and VGG-16 and VGG-16 without applying weightage to imbalance dataset respectively, model B and model D also work well in terms of positive sample prediction but have very low specificity while model C has low sensitivity. The confusion matrix of all models is provided in Figure 11. The performance metrics of all models are given in Table 1. The limitation factor of model is the size of test and train dataset.

Figure 11: Confusion Matrix of selected models.
The formulae used to calculate Precision, Recall, Specificity and F1 score is provided as Equation 1, Equation 2, and Equation3 and Equation 4 respectively.
Precision (P)= TP/(TP+FP) … Equation 1
Recall (R)=TP/(TP+FN) … Equation 2
Specificity= TN/(TN+FP) … Equation 3
F-Score =2*P*R/(P+R) … Equation 4
Assay Senstivity and Specificity
As we processed 20 positive samples with different Ct values and 18 negative samples, it was observed that from 20 samples, 17 samples produced positive results while 3 samples gave negative results in our assay. On the other hand, 18 PCR negative samples were also processed for the validation of the in-house developed assay, from which 16 samples showed true negative while 2 samples showed false positive results. So, the sensitivity and specificity of the assay was calculated to be 85% and 88% respectively. Final results were evaluated using light microscopy and digital microscopy images.
Discussion
Since the beginning of the pandemic, it was paramount to develop rapid testing methods to detect COVID-19 so that containment measures could be taken and timely diagnosis and treatment of the infection could be done. One of those rapid testing approaches is latex agglutination to detect either antibodies or antigens for the diagnosis of COVID-19. In this study, we developed an immunological method to detect SARS-COV-2 based on its binding with the antibodies which are affinity-purified and passively adsorbed onto the polystyrene latex beads. We used PCR-positive samples to assess the working of our assay. During the study, we observed that the patients’ samples revealed a distinct band pattern with homogenous agglutination as seen in the recent pilot study conducted in 2021 [13] while the negative samples and blank did not show any type of agglutination.
We also performed the microscopic analysis of the slides at 100X magnification, contrary to the study in which visualization was done on 4X magnification which revealed dark blue dots in positive samples [14] while in our study, microscopic investigations showed a white discrete patchy appearance in positive samples corresponding to the presence of virus and its interaction with the antibodies on the beads. Moreover, it was also seen that the discrete white patches were seen in larger quantities in samples with high Ct-values indicating high viral load while the opposite was observed in low Ct-value samples and in negative samples.
Furthermore, we diluted the positive samples initially with three dilutions i.e. 1:1, 1:2 and 1:10 in which we evaluated that 1:10 dilution did not show any results while 1:1 and 1:2 showed satisfactory results with patchy appearance, so we further proceeded with the two dilutions only which showed prompt outcomes with respective to Ct-values of the samples. It was interpreted from these findings that antibodies onto the beads must be saturated with an adequate number of antigens in the sample to produce distinct and differentiable results. To further make this technique handier, the digital microscope was used which is comparatively cheap and easy to access as compared to the light microscope having a magnification of 1600X. Positive samples were seen to have a bright, shiny crystal-like appearance which was interpreted as the agglutinated area where the binding of viral antigens with antibodies took place while there was no such thing seen in negative samples and blank as they just gave a dark blue hollow appearance indicating the presence of dark blue dyed latex beads without any presence of antigen.
To validate the findings on light and digital microscope, another microscopic analysis was done i.e. Electron microscopy in which agglutination in positive samples was seen as protrusions or hump formation. Beads were aggregated so compactly that it gave an appearance similar to a hump which was absent in negative samples. In negative samples, only beads with conjugated antibodies on their surface were seen under an electron microscope.
Furthermore, to give the broad application of the developed assay an application of artificial intelligence is implied to images obtained from Light microscopy to make the tool usable to even those that have little knowledge of interpreting Light microscopy images. A convolutional Neural Network (CNN) has been constructed to classify the images as positive and negative. CNN is considered as successful machine-learning approach in terms of image classification. The concept of the CNN network is driven by animal neural cortex [15]. To date, CNN has been successfully applied for bio-medical image classification like in cancer histology sample analysis and in microscopic image classification of different cell types [11,16]. Model E performs well as compared to other models and has been shortlisted. Nevertheless, this is the first step towards incorporation of AI in agglutination reaction which will be further enhanced in terms of specificity and sensitivity by increasing the sample size for both training and testing dataset.
Added to that, we further aim to refine this rapid test by creating a digital microscope image library of SARS-COV-2 positive and negative latex agglutination tests and using different Artificial Intelligence (AI) tools to create the software. The combination of a latex agglutination test, a digital microscope, and AI-based software in a smartphone device may be utilized as a futuristic approach to accurately detect the SARS-COV-2 infection in diseased patients.
This assay has some limitations which include the size of the agglutinates and the number of samples tested. As the clumps are very small, it is hard to observe them from the naked eye so, further studies can be done to improve this phenomenon by attaching either a chromophore or a fluorophore to the beads or by using functionalized latex beads for antibody conjugation which can promote better visualization by producing stabilized protein complex with the beads. Another limitation is the restricted number of samples which can be increased for further quality control and validation studies in the future.
Conclusion
In this study, we have developed a convenient testing method based primarily on a microscopic approach for the diagnosis of COVID-19 by employing the latex beads agglutination technique. We found that the interaction between the whole virus with its corresponding antibodies results in the formation of discrete-white patches and crystal-like shiny aggregates which can be easily seen through a light microscope as well as a digital microscope on which clumping of beads was seen in positive samples indicating the binding of antibodies to the virus. Furthermore, a CNN model based on light microscopy images processing was constructed to amplify the agglutination test as the point of care testing assay. To the best of our knowledge, this is the first ever study demonstrating the potential of a latex agglutination assay as a rapid diagnostic test by coupling it with a hand-held digital microscope and an AI tool for the detection of SARS-COV-2.
Author Statements
Authors’ Contributions
NS performed all the experimentation and wrote the manuscript, KI interpreted the results and supervised the study along with reviewing this manuscript, RB designed the immunization plan and slide agglutination assay, FJ designed the AI-based image library, NA, MS, OQ and FA provided their expertise and assistance in microscopy and PCR samples’ validation, AT played a major role in adsorption strategy development, SK oversaw the whole study and provided his intellectual input.
Declaration of Interest
The authors affirm that the study was carried out without any commercial or financial associations that could be seen as a possible conflict of interest.
Data Availability Statement
The data that supports the findings of the study are available from the corresponding author, [KI], upon reasonable request.
References
- Li H, Liu SM, Yu XH, Tang SL, Tang CK. Coronavirus disease 2019 (COVID-19): current status and future perspectives. Int J Antimicrob Agents. 2020; 55: 105951.
- Peng M. Outbreak of COVID-19: an emerging global pandemic threat. Biomed Pharmacother. 2020; 129: 110499.
- Baj J, Karakula-Juchnowicz H, Teresinski G, Buszewicz G, Ciesielka M, Sitarz R, et al. COVID-19: specific and non-specific clinical manifestations and symptoms: the current state of knowledge. J Clin Med. 2020; 9: 1753.
- Jarvis MC. Aerosol transmission of SARS-CoV-2: physical principles and implications. Front Public Health. 2020; 8: 590041.
- Tahamtan A, Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev Mol Diagn. 2020; 20: 453-4.
- Rasmi Y, Li X, Khan J, Ozer T, Choi JR. Emerging point-of-care biosensors for rapid diagnosis of COVID-19: current progress, challenges, and future prospects. Anal Bioanal Chem. 2021; 413: 4137-59.
- Habli Z, et al. COVID-19 in-vitro diagnostics: state-of-the-art and challenges for rapid, scalable, and high-accuracy screening. Front Bioeng Biotechnol. 2021; 8: 1562.
- Silveira-Gomes F, Sarmento DN, Pinto TM, Pimentel RF, Nepomuceno LB, Espírito Santo EP, et al. Development and evaluation of a latex agglutination test for the serodiagnosis of paracoccidioidomycosis. Clin Vaccine Immunol. 2011; 18: 604-8.
- Elias A, Aptel I, Huc B, Chalé JJ, Nguyen F, Cambus JP, et al. D-dimer test and diagnosis of deep vein thrombosis: a comparative study of 7 assays. Thromb Haemost. 1996; 76: 518-22.
- Bangs L. TechNote# 204: adsorption to microspheres. Fishers: Bangs Laboratories. 1999.
- Oei RW, Hou G, Liu F, Zhong J, Zhang J, An Z, et al. Convolutional neural network for cell classification using microscope images of intracellular actin networks. PLOS ONE. 2019; 14: e0213626.
- Morid MA, Borjali A, Del Fiol G. A scoping review of transfer learning research on medical image analysis using ImageNet. Comput Biol Med. 2021; 128: 104115.
- Cariaga-Martínez A, GK, AA. R, pilot assessment of a rapid test for the detection of COVID-19 disease by using latex agglutination. Austin J Clin Case Rep. 2021; 8: 1224.
- Elliff J. Development of a rapid diagnostic test for the detection of antibodies or antigens to coronavirus (COVID-19). Fields: Journal of Huddersfield Student Research. 2022; 8: 1-23.
- Hubel DH, Wiesel TN. Receptive fields and functional architecture of monkey striate cortex. J Physiol. 1968; 195: 215-43.
- Rakhlin A, et al. Deep convolutional neural networks for breast cancer histology image analysis. In: Image Anal Recognit: 15th International Conference, ICIAR 2018, Póvoa de Varzim, Portugal, Jun 27-29, 2018, Proceedings. 2018; 15.