
Research Article
Austin J Biosens & Bioelectron. 2023; 8(1): 1046.
Network Analysis of Differential Expression of ANLN and Its Interacting Proteins in Esophageal Squamous Cell Carcinoma
Hong Sun1,2; Yufei Cao1,2; Bingli Wu1,2; Beibei Tong1,2; Huayan Zou1,2; Liyan Xu2; Enmin Li1-3*
¹Department of Biochemistry and Molecular Biology, Shantou University Medical College, Shantou 515041, China
²Key Laboratory of Molecular Biology in High Cancer Incidence Coastal Chao Shan Area of Guangdong Higher Education Institutes, Shantou University Medical College, Shantou 515041, China
³Shantou Academy Medical Sciences, Shantou 515041, China
*Corresponding author: Enmin LiDepartment of Biochemistry and Molecular Biology, Shantou University Medical College, Shantou 515041, China. Email: nmli@stu.edu.cn
Received: August 31, 2023 Accepted: September 25, 2023 Published: October 02, 2023
Abstract
Anillin (ANLN) is an actin binding protein, which was originally extracted from Drosophila melanogaster embryos. As a key regulatory factor in cytokinesis, ANLN is highly expressed in various tumors, leading to abnormal cell division and promoting the proliferation, migration, and invasion of cancer cells. At present, the role and regulatory mechanism of ANLN in human Esophageal Squamous Cell Carcinoma (ESCC) are not fully understood. The purpose of this study is to construct a Protein-Protein Interaction Network (PPIN) to reveal the characteristics of ANLN and its interacting proteins, by the integration of their expression in ESCC. The differentially expressed ANLN and its interacting proteins in ESCC were identified from our previous RNA-seq data. By constructing a specific PPI network, it was found that many differentially expressed genes/proteins may interact with ANLN. Multiple enrichment pathways of ANLN and its differentially expressed genes were explained by functional enrichment analysis, Gene Ontology (GO) analysis and KEGG pathway analysis. In addition, it is revealed that ANLN, ECT2, ACADM and PPP1R9A play an important role in the occurrence and development of ESCC, and they are proposed as new prognostic factors for ESCC. These bioinformatics analyses provide a comprehensive perspective for the role of ANLN in ESCC.
Keywords: ANLN and its interacting proteins; Differential expression; Protein-protein interaction network; Esophageal squamous cell carcinoma
Introduction
In the human genome, ANLN is located on the human chromosome 7p14.2 and can encode 1124 amino acids with a theoretical molecular weight of 124 kDa [1]. ANLN is a scaffold protein with several unique domains. Its N-terminal contains several domains that bind to the contractile ring proteins, and its C-terminal contains RBD, C2 and PH domains that bind to plasma membrane [2]. ANLN is an actin binding protein, which was originally extracted from the embryos of Drosophila melanogaster [3]. ANLN plays a role in cell growth, migration and cytokinesis [4]. As a highly conserved protein, ANLN can regulate the process of cell division by interacting with different proteins such as F-actin, myosin II and septins [5]. During cytokinesis, ANLN is concentrated in the contractile ring to provide contractile force and mediate the separation of daughter cells [6]. ANLN improves contraction efficiency by directly binding to phosphorylated myosin, or by coordinating the effective interaction between F-actin and Non-muscle Myosin II (NM II) [7,8]. ANLN combines with septins, an important regulator of cell division and mechanical transduction, to regulate cell proliferation, differentiation and migration [9,10]. Several studies have found that ANLN plays a crucial role in promoting tumor cell proliferation, and its absence can inhibit tumor cell division [11,12]. In addition, the promoting effect of ANLN on the growth of gastric cancer cells in vivo has been confirmed by mouse models [13]. The high expression level of ANLN in breast cancer cells is related to the poor prognosis of patients with breast cancer [14]. ANLN is related to T lymphocyte infiltration in pancreatic cancer and has certain potential value in immunotherapy [15]. ANLN enhances the metastasis of lung adenocarcinoma by promoting epithelial-mesenchymal transformation [16]. Targeting USP10-ANLN axis can effectively inhibit the progression of cell cycle in ESCC [17]. Overall, the high expression of ANLN leads to abnormal cell division, thereby promoting the proliferation, migration, and invasion of cancer cells [18-23].
ESCC is a common malignant tumor of digestive system with a high morbidity and mortality. According to the global cancer statistics in 2020, the incidence of ESCC ranks seventh and its
mortality ranks sixth among all malignant tumors [24]. ESCC is the most common subtype of esophageal cancer worldwide, with a high prevalence in East, East and South Africa and southern Europe [25], and its five-year survival rate is 15% - 25% [24], which is a serious threat to human health [26]. China is a country with a high incidence of ESCC, accounting for more than half of the world's morbidity and deaths [27]. ESCC is the fourth leading cause of death of malignant tumors in China, leading 375,000 patients die of ESCC [28]. With the continuous accumulation of biological knowledge and various omics data, protein-protein interaction data are becoming more and more abundant, which makes it possible for them to be used to construct gene regulatory networks for tumors and other diseases. PPI network can provide a powerful working platform for revealing the pathogenesis of complex diseases and drug development [29-32]. In this study, we collected the interaction proteins of human ANLN, constructed a PPI network containing differentially expressed ANLN interacting proteins in ESCC, and analyzed its clinical significance. These data contribute to reveal the role of ANLN in ESCC cells and provide an important molecular basis for studying the regulatory mechanism of ANLN in ESCC.
Materials and Methods
Expression of ANLN and its Interacting Proteins in ESCC
The gene set of ANLN and its interacting proteins was collected from NCBI (https://www.ncbi.nlm.nih.gov/), HPRD (http://www.hprd.org/) (Release 9) and BioGRID (http://thebiogrid.org/) (Release 4.4.209). In our previous high-throughput RNA-seq data, we have screened the expression trend of ANLN and its interacting proteins in ESCC from 15 pairs of ESCC clinical samples using the same stringent criteria. (Foldchange >2 or <0.5, FDR value < 0.05) [33]. The differentially expressed genes/proteins which interacts with ANLN are used to the construction of PPI networks.
Construction of the ANLN PPI Network
The up to date human protein-protein interaction data sets were downloaded and collated from HPRD (http://www.hprd.org/) (Release 9) and BioGRID (http://thebiogrid.org/) (Release 4.4.209) respectively. These two data sets contain low-throughput and high-throughput experimental results collected from public references [34,35]. Based on the above data, a complete human protein-protein interaction network was constructed by Cytoscape software, and the duplicated edges and self-loops in the network were deleted through the "Network Modification" menu to avoid chaos in the calculation of topology parameters of the PPI subnetwork [36,37]. This newly generated network contains 21,678 unique proteins and 790,776 pairs of interactions, which was considered as parent PPI networks. It is well known that up-regulated and down-regulated genes or proteins play an important role in tumorigenesis [38]. In order to highlight their importance, according to the steps we described earlier, ANLN and its interacting proteins with differential expression in ESCC are generated by Cytoscape software to generate specific PPIN [39]. Firstly, the ANLN related dysregulated GENE IDs (official gene symbol) were listed in a text file and mapped to the parental PPI network imported into Cytoscape by the menu of "Select→Nodes→From ID List File". Secondly, the first level interactions between ANLN and their neighbor were detected by menus of "Select→Nodes→First Neighbors of Selected Nodes" and "New→Network→From Selected Nodes, All Edges" to obtain the ANLN PPI network.
Topology Analysis of ANLN PPINs
The Network Analyzer plugin in Cytoscape software was used to analyze the topology parameters of ANLN PPI network, and several topology metrics were calculated, such as clustering coefficient, degree distributions, neighborhood connectivity, topological coefficient and so on, to deeply reveal the organization and structure of complex networks [40]. The node degree to the number of edges associated with the node, or the number of connections, and the node here refers to the protein. Obviously, the more protein nodes in the network, the greater the degree of the node, and the power distribution of the node is the most significance. The network topology feature analysis method and its important parameters were described in our previous work [39]. The degree distribution of the network satisfies the following relation, P(K)=n(K)/N, that is, the ratio of all nodes with k to the total number of nodes in the network. If there are n nodes in a network and the value of n (K) nodes is K, then P(K)=n(K)/N. The degree distribution is the whole distribution of P(K). Network Analyzer can also draw the power distribution curve y=߷xa and calculate the R² value of the fitting degree of the reaction power distribution curve. The closer the R² value is to 1, the higher the curve fitting degree is. In addition, Network Analyzer can also calculate several other important topology parameters at the same time, including the shortest path length, compactness center and so on.
Subcellular Layer of ANLN PPIN
The subcellular localization information of each protein in the ANLN PPI network were downloaded from the HPRD database, and then imported them into the network as the attributes of the nodes. With the cerebral plugin in Cytoscape, nodes were rematched to different subcellular locations, which did not change the interaction between nodes [41].
Functional Enrichment Analysis of Differentially Expressed Genes Related to ANLN
In order to further understand the functional relationship between ANLN and its interacting proteins, the functional annotation map module in Bioinformatics Database (DAVID) (https://david.ncifcrf.gov/) was used to analyze and annotate the genes related to the up-regulated or down-regulated expression of ANLN in ESCC. The results with statistical significance (P<0.001, FDR<0.05) were screened and visualized by Enrich map plugin in Cytoscape. Functional classification includes Gene Ontology (GO), INTERPRO, KEGG, SMART and other annotation categories [42].
GO Enrichment Analysis
GO Tree is an important part of WebGestalt (web-based gene set analysis toolkit) [43,44]. It uses GO Directed Acyclic Graph (DAG) to construct and visualize gene sets, including extensible trees, bar graphs of selected annotation layers and enriched DAGs. Among them, DAG is the most intuitive form of GO enrichment analysis, which mainly reflects the hierarchical relationship between the superior and subordinate of GO terms and the degree of enrichment, and it is used to visualize GO categories with a rich number of genes identified by statistical modules. The darker the color in the graph, the more significant the richness, while colorless means that the richness is not significant. In addition, the histogram can also be used to count the enrichment results.
KEGG Pathway Enrichment Analysis
One of the most crucial tasks in biological research is to determine the molecular pathway in which proteins are involved. WebGestalt can display the name of each KEGG pathway associated with the gene set studied, all genes involved in the pathway, and the corresponding Entrez ID [43,45]. In addition, the KEGG table not only provides a P value that reflects the importance of each pathway, but also links to the KEGG map, where the genes in the gene set are marked in red.
Survival Analysis of ANLN-Related Differentially Expressed Genes in ESCC
The expression data (GSE53625) was downloaded from the GEO database, which contains 179 pair’s samples of ESCC. Those cases with survival of <3 months were excluded, leaving only 175 cases of ESCC. According to the differential gene expression level, the best segmentation point was found by X-Tile (verison 3.6.1) software, and the ESCC patients were divided into high expression group and low expression group. Kaplan-Meier and Log-rank tests were used to analyze the survival of the patients, and draw the survival curve by GraphPad Prism 5 [46,47].
Results
The Differential Expression of ANLN and its Interacting Proteins in ESCC
In our previous study, we performed high-throughput RNA sequencing (RNA-seq) in 15 pairs of ESCC clinical samples to target the key function lncRNAs that regulates expression of downstream protein-coding genes in ESCC [33]. In order to detect the expression pattern of ANLN and its interacting proteins in ESCC, we analyzed their expression level in RNA-seq data with the fold change >2 or <0.5 (FDR<0.05). In this data, there are 192 differentially expressed genes, of which 155 genes were up-regulated, fold change ranging from 2.01 to 12.1, and 37 genes were down-regulated, fold change ranging from 0.043 to 0.493 (Figure 1).
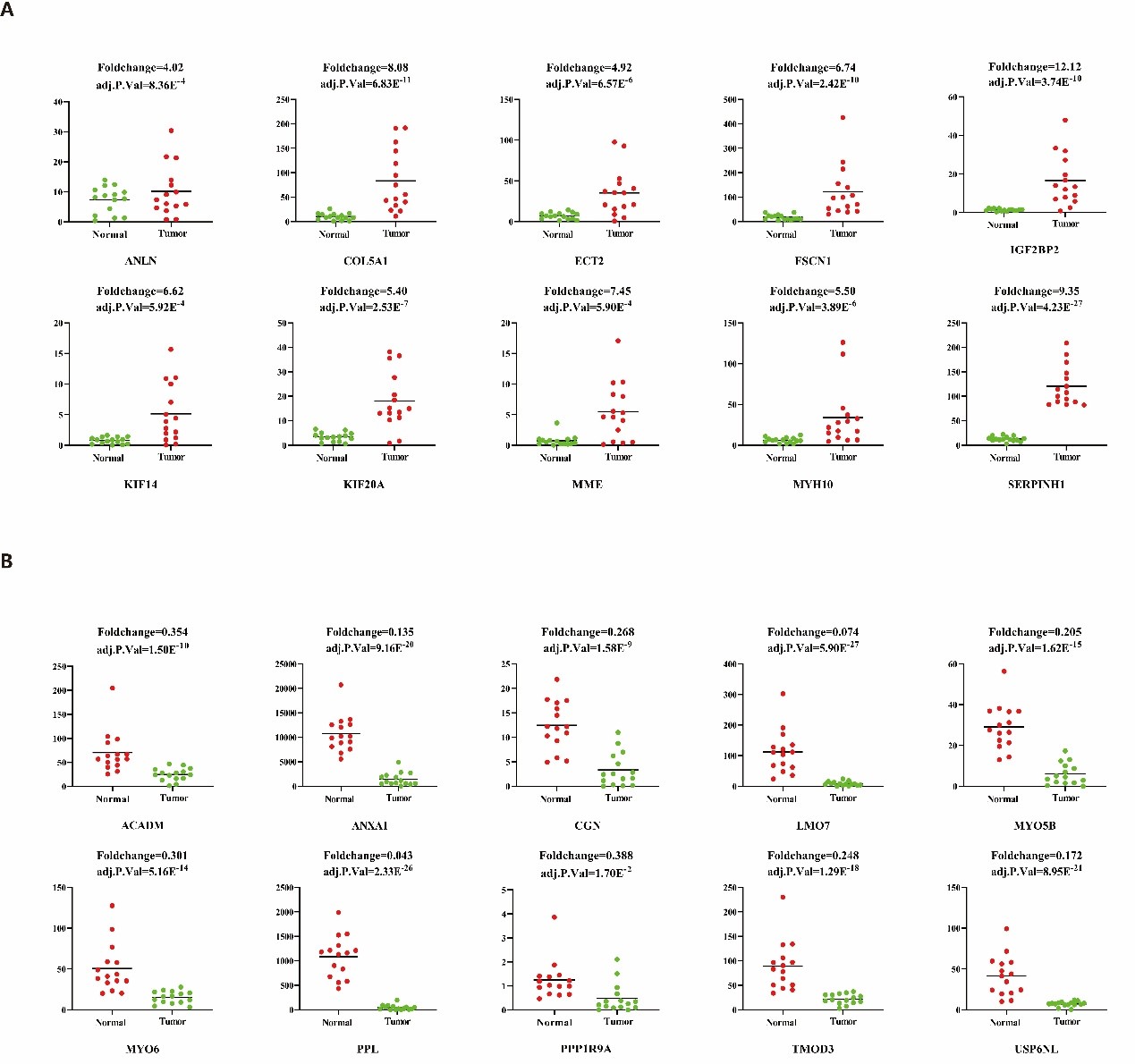
Figure 1: Significant dysregulated ANLN and its interacting proteins in ESCC. These genes were identified from previous RNA-seq data, with red indicating the up-regulated and green indicating the down-regulated. (A) Ten up-regulated ANLN and its related genes in ESCC. (B) Ten down-regulated ANLN-related genes in ESCC.
The Construction of ANLN PPIN
In order to understand the biological function and significance of ANLN, we constructed a panoramic picture of the cellular biological activity of ANLN and screened its interacting proteins to reveal their role in ESCC. A total of 192 differentially expressed ANLN-related genes are mapped to parent interaction networks as key hub nodes to form a sub-network containing all their interactions. It consists of 1,079 nodes and 40,162 edges and is called a differential ANLN interacting network (Figure 2A). To facilitate a clearer understanding of the network, we created another concise network of 192 aberrant genes and excluded the interaction between the other proteins, leaving only their direct interaction with ANLN (Figure 2B). The results show that hundreds of other proteins are linked together, and these proteins can directly or indirectly form protein complexes with different functions with ANLN. As a result, the biological consequences may be affected by their differential expression.
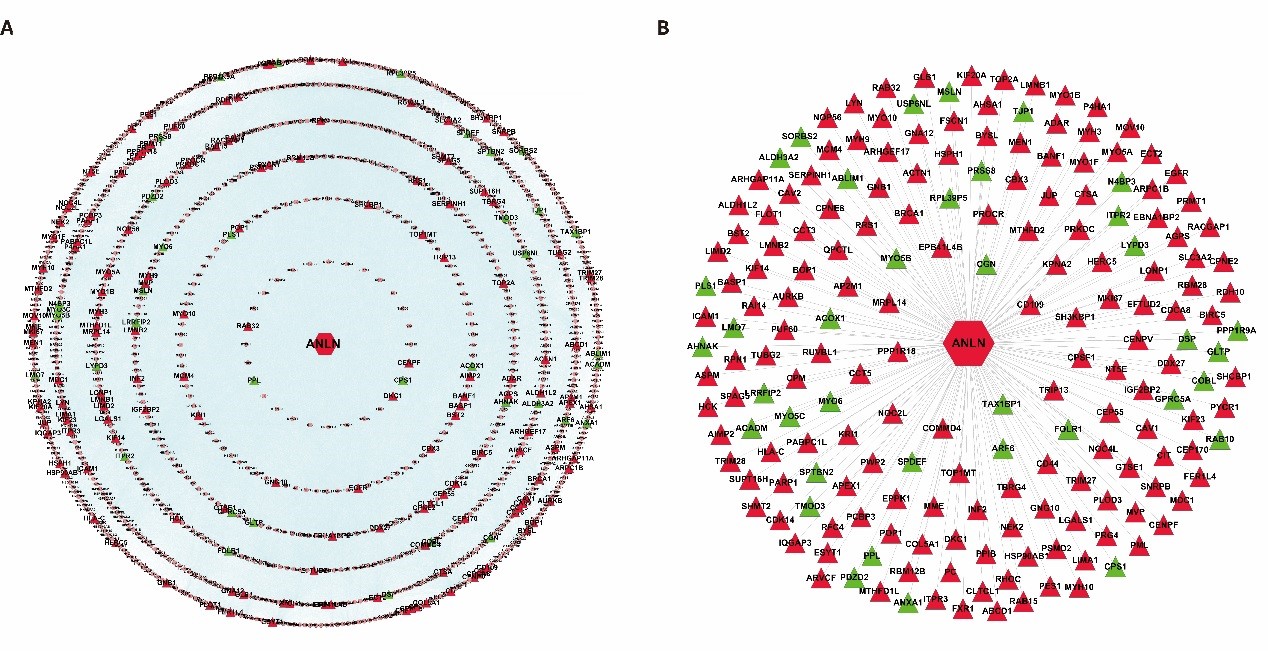
Figure 2: Two different ANLN interaction networks. (A) The differentially expressed ANLN PPIN with 155 up-regulated genes and 37 down-regulated genes as the central node, including the interaction between proteins. (B) Compared with A, it looks clearer and more concise. It is a more concise PPI sub network directly constructed by ANLN and its interacting proteins. Red indicates up-regulation and green means down-regulation.
Topological Properties of ANLN PPI Network
Different types of networks have different topological characteristics. Protein-protein interaction network is a kind of biological network, and its node degree power distribution is one of the most important characteristics [48,49]. As shown in Figure. 3A, the node degree distribution of the network obeys the power distribution, and the heavy-tailed form is y=74.816x-0.716 and R²=0.654. In many other co-expressed protein-protein interaction networks, R² is also higher than 0.6 [50,51]. We also tried to analyze the shortest path length of ANLN PPIN (Figure3B). The results show that the shortest path length between the two proteins in the network is mainly concentrated in 2, indicating that ANLN interacting protein network is closely related to each other. It also shows that these proteins are directly or indirectly associated with another protein. The key protein nodes are closely linked, and the network composed of them is more stable [52]. As shown in Figure 3C, the domain connectivity of ANLN PPIN, that is, the average connectivity between adjacent proteins approximately shows a slow linear downward trend. This shows that there are not many highly connected proteins associated with ANLN in the network, and they happen to be the key proteins in the network.
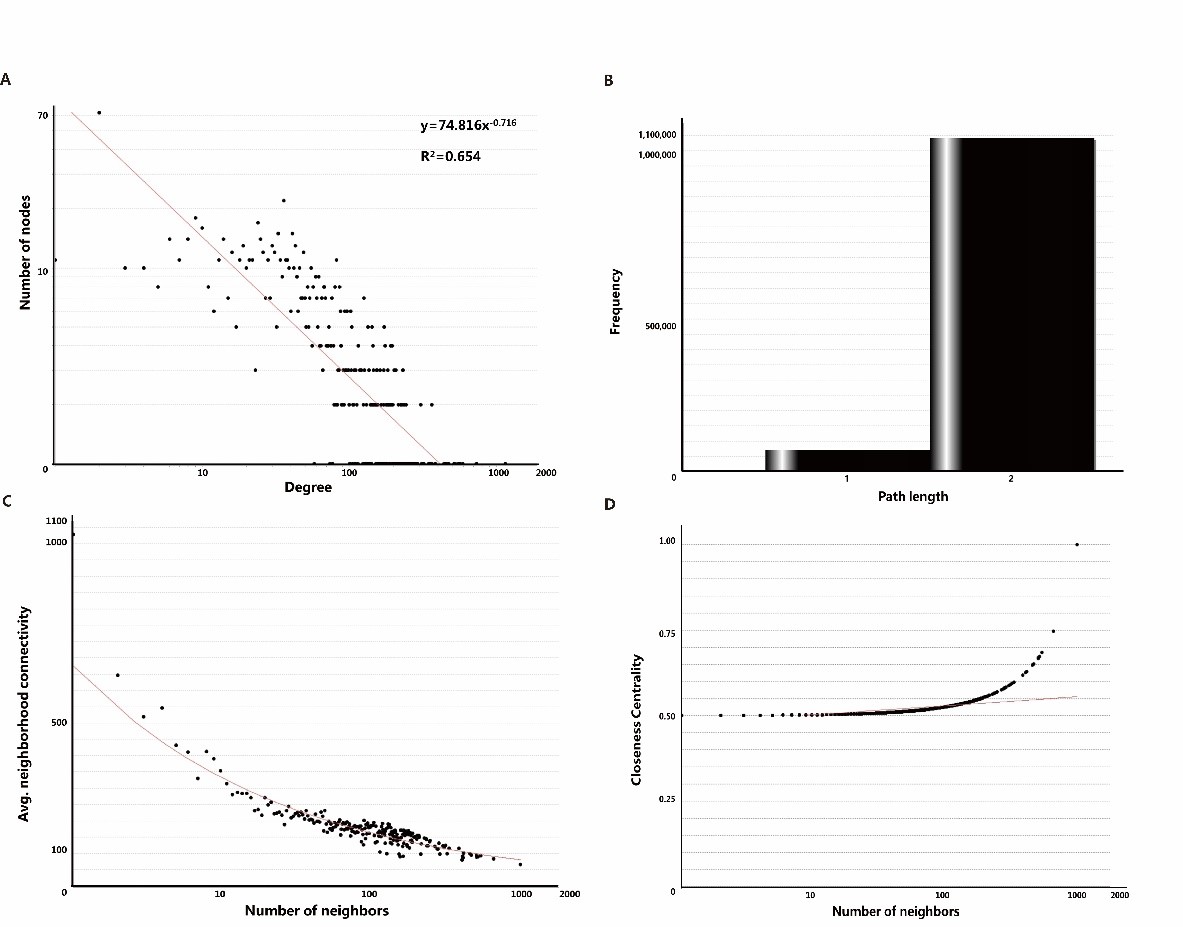
Figure 3: The main topological properties of differentially expressed ANLN PPIN. (A) Degree distribution of the ANLN PPIN. (B) Shortest path length of the ANLN PPIN. (C) Neighbor connectivity of the ANLN PPIN. (D) Closeness centrality of the ANLN PPIN.
Compact centrality measures the distance between the node and other nodes in the network. If the shortest distance between the node and other nodes on the way is very small, then the tight centrality of the node is high. This is also related to the length of the shortest path between nodes (Figure 3D). These curves show that there is a close relationship between several proteins.
Subcellular Layer Analysis of ANLN PPIN
Subcellular localization can locate a protein in a specific location in the cell, and mature proteins can play a stable biological function in specific subcellular organelles [53,54]. In PPI network, nodes can be distributed through cerebral plugin, and the interaction between nodes will not change due to the change of their subcellular position. ANLN PPIN is divided into 10 levels according to percentage: secreted (1.11%), membrane (10.57%), membrane/nucleus (0.28%), cytoskeleton (1.11%), cytoskeleton/cytoplasm (0.37%), cytoplasm (20.39%), secreted/nucleus (0.19%), cytoskeleton/nucleus (1.11%), cytoplasm/nucleus (27.15%), nucleus (17.33%) and uncertainty (20.57%) (Figure 4A). These results not only indicate that some ANLN-related proteins interact with them on multiple subcellular organelle, but also show that ANLN PPI networks can transmit intracellular signals or complexes from outside to inside through other specific pathways, and finally achieve the direct distribution of nuclear ANLN-related differentially expressed proteins. The protein-protein interaction network also clearly reflects their subcellular localization, as shown in Figure 4B.

Figure 4: Subcellular layers of ANLN PPINs in ESCC. (A) Subcellular localization distribution of the differential ANLN PPIN. (B) Subcellular localization distribution of the concise ANLN PPIN.
Functional Enrichment Analysis of ANLN differentially Expressed Protein Network
In order to further understand the potential function and significance of ANLN and its related differential proteins, the ANLN differential protein network was annotated through David bioinformatics database, and then visualized using the Enrichment Map plugin in Cytoscape. According to the classification of functional tagging terms, the ANLN differential protein networks in ESCC were divided into several categories, such as GO, INTERPRO, KEGG, SMART and so on. As shown in Figure. 5, each node has a dedicated comment that describes its functional characteristics. Different types of categories are represented by different colors. The overlap coefficient between these categories (the overlap coefficient cutoff value 0.9) is defined as the width of the edge, and the more genes are shared between the two nodes, the wider the edge is.
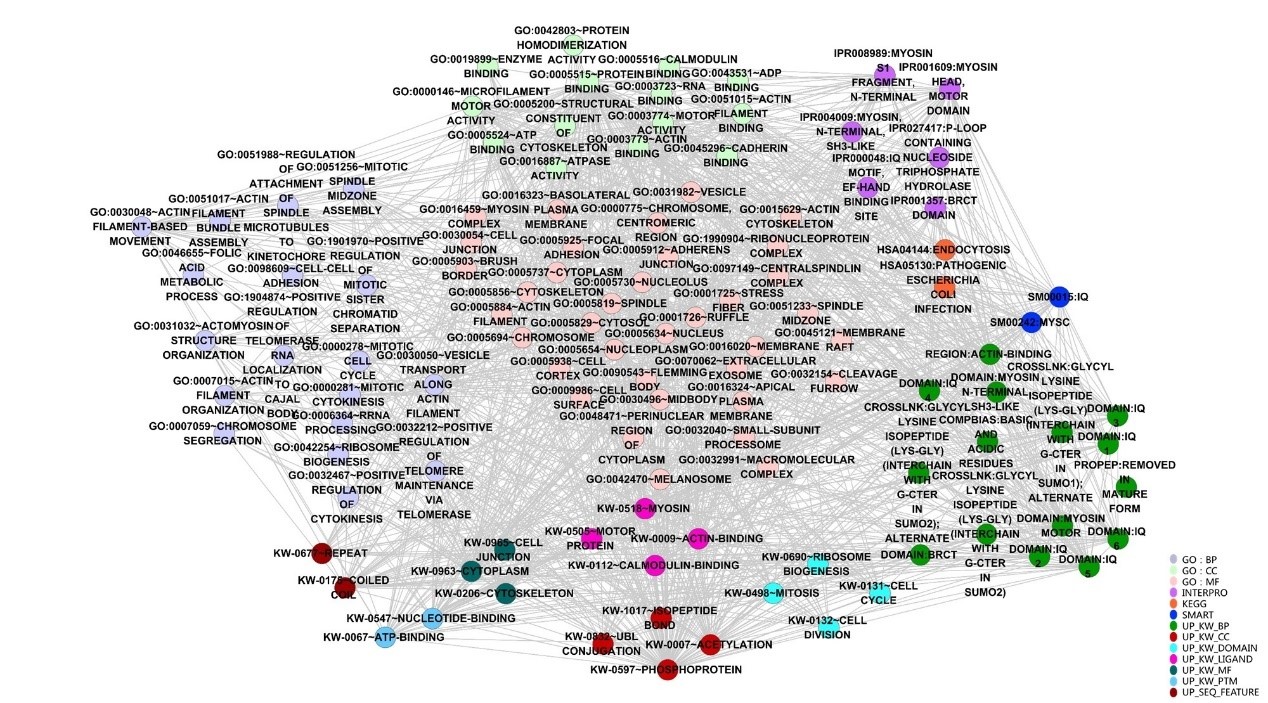
Figure 5: Visualization of functional categories of the differential ANLN PPIN.
GO Analysis of ANLN Network
WebGestalt, a functional enrichment analysis web tool, was be used for GO enrichment analysis of ANLN network. Compared with the GO annotation, we can find many known functions of ANLN and its interacting proteins as well as unexplored functions (Supplementary Table 1). The biological process is represented by the red bar chart, the cellular composition is represented by the green bar chart, and the molecular function is represented by the blue bar chart, respectively. The longer the bar, the closer the function is to the ANLN (Figure 6A). As shown in Figure. 6B, ANLN was indirectly involved in the biological processes of actin filament organization (GO: 0007015), regulation of actin filament-based process (GO: 0032970), regulation of cytoskeleton organization (GO: 0051493), actin filament-based movement (GO: 0030048). At the molecular functional, ANLN was associated with cell adhesion molecule binding (GO: 0050839) and actin binding (GO: 0003779). In the category of cell composition, we found that ANLN was closely connected with these functions, such as actin cytoskeleton (GO: 0015629), midbody (GO: 0030496), cell division site (GO: 0032153), cytoplasmic region (GO: 0099568).
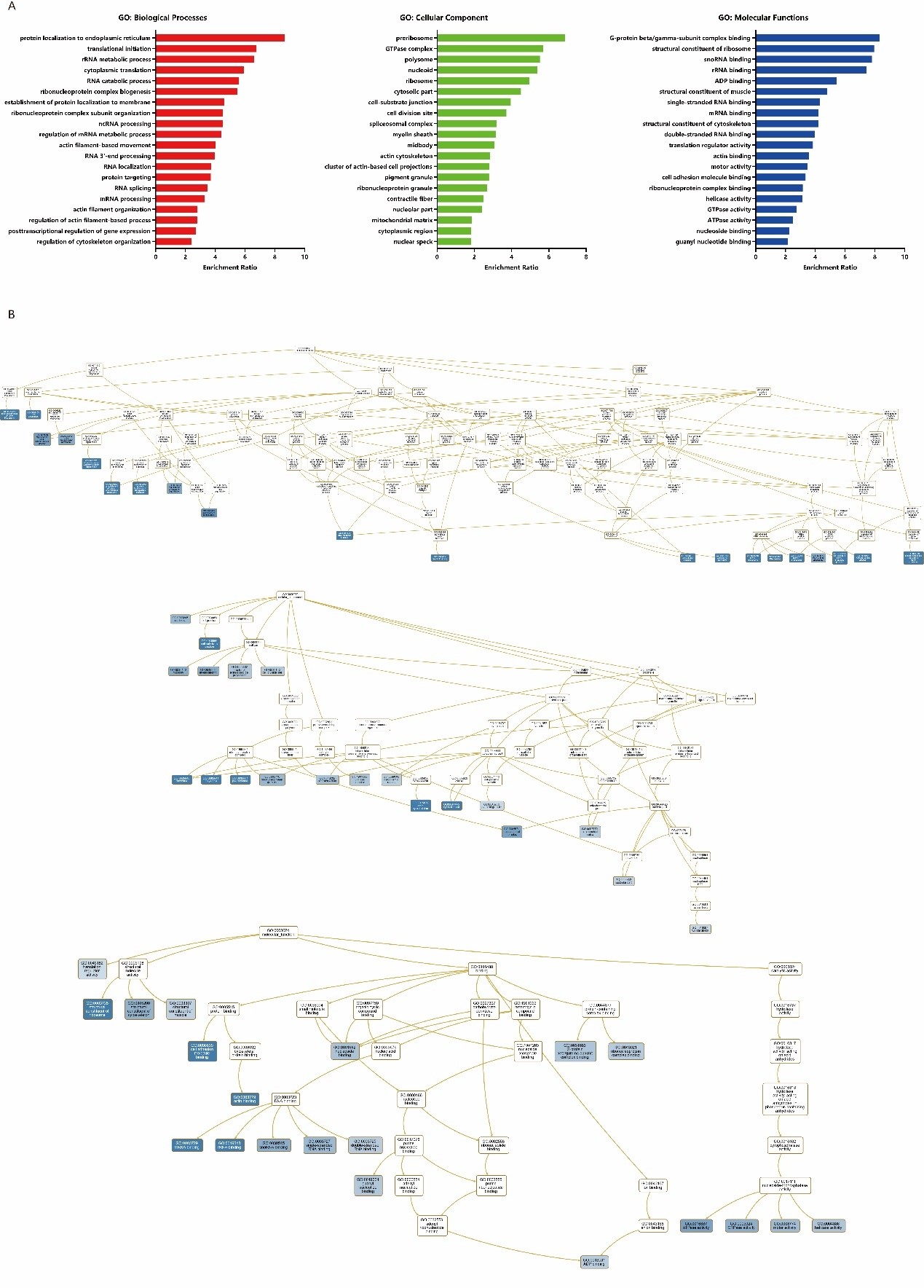
Figure 6: GO enrichment analysis of ANLN network in ESCC. (A) The bar chart for GO enrichment analysis. Different colors
represent different GO categories. (B) DAG for GO enrichment analysis. The darker the blue, the higher the enrichment ratio.
KEGG Pathway Enrichment Analysis
ANLN and its interacting proteins are enriched to the associated signaling pathways on KEGG pathway. There are 41 pathways had been enriched in this study (Figure 7, Supplementary Table 2). More than 40 ANLN related genes are involved in these pathways, including ribosome (hsa03010), spliceosome (hsa03040), and regulation of actin cytoskeleton (hsa04810), endocytosis (hsa04144). It is suggested that the protein ANLN interaction network is closely related to these pathways.
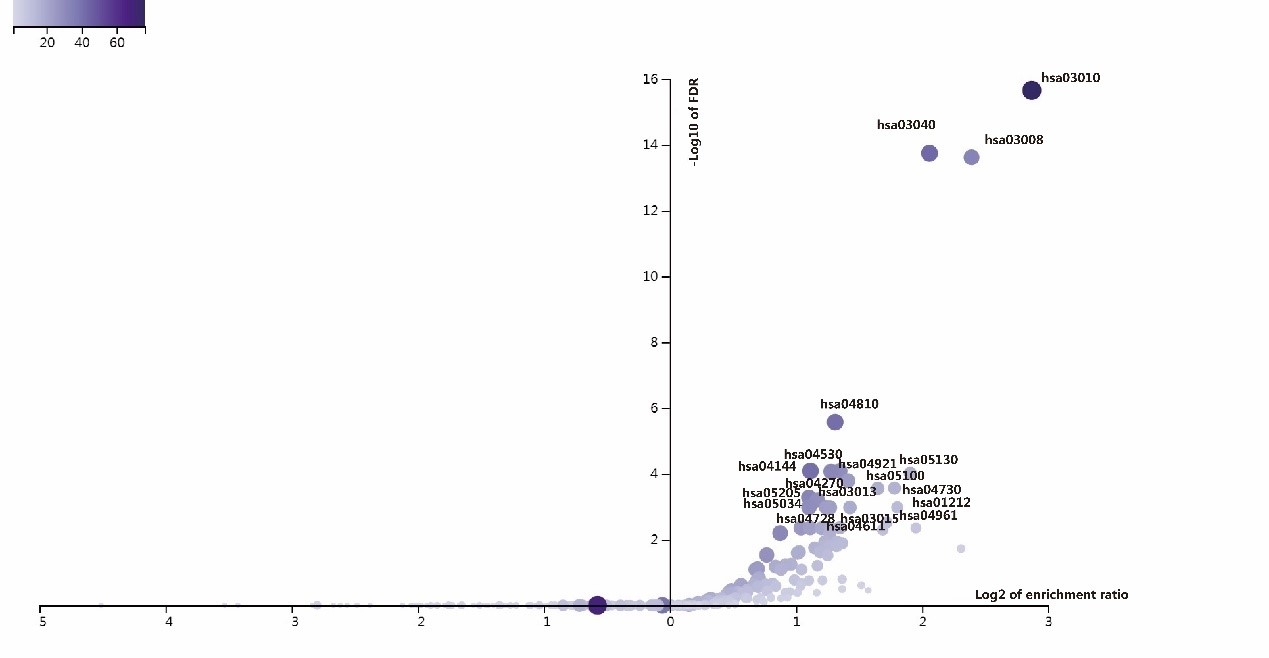
Figure 7: The volcanic map of enrichment of KEGG pathway in ANLN PPI network.
ANLN and its Interacting Proteins Correlate with the Survival of ESCC Patients
In order to verify the clinical significance of ANLN and its interacting proteins in ESCC, we used GSE53625 expression profile data for survival analysis. After optimized typing by X-Tile software, a total of 4 ANLN and its related differential expression genes were considered to be significantly correlated with the survival time of patients with ESCC (Figure 8). The survival rate of patients with the high expression of genes (ACADM, ANLN, ECT2 and PPP1R9A) were generally worse than that of patients with the low expression one.
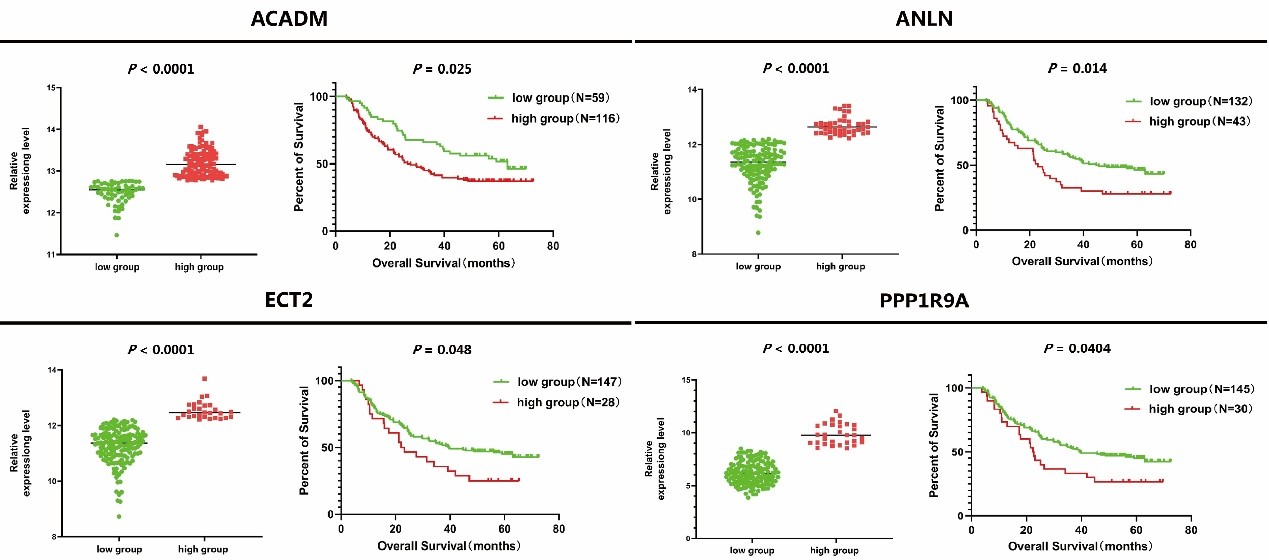
Figure 8: The relationship between expression level of ANLN and its related differentially genes and survival rate in patients with ESCC.
Discussion
ESCC is a common malignant tumor of digestive tract. According to the global cancer statistics in 2020, the morbidity and mortality of ESCC are among the top ten among all malignant tumors. There are two common histological types of esophageal cancer, Esophageal Adenocarcinoma (EAC) and Esophageal Squamous Cell Carcinoma (ESCC). EAC is the main subtype of esophageal cancer in Europe and the United States and other western society. Since 1984, the incidence of EAC in developed countries has gradually increased [55]. The prevalence of ESCC is high in East, East and South Africa and Southern Europe [25]. China is in a high-risk area of ESCC, and it is a country with a high incidence of ESCC, with more than half of the annual morbidity and deaths in the world [24,27]. Due to the difficulty of early diagnosis, most patients with ESCC are in the middle and late stage of the disease at the time of diagnosis, which is the main reason for their low overall 5-year survival rate [56]. In order to improve the survival of patients with ESCC, it is an urgent problem to find biomarkers and treatment targets for diagnosis and prognosis. Proteins interact with each other to form macromolecular structures and enzyme complexes, which are critical for cell survival and growth. The imbalance of protein-protein interaction leads to cellular functional defects and various diseases [57]. Therefore, PPI networks can provide new insights into the molecular mechanisms of specific genes/proteins or diseases [58,59]. A large number of studies have shown that comprehensive analysis of gene expression and PPIN can provide in-depth insights into the molecular mechanisms of diseases or specific genes [60,61]. This provides research direction for the discovery of molecular markers for cancer diagnosis, and provides a theoretical basis for revealing the pathogenesis of cancer and drug development.
We constructed a specific protein-protein interaction network using 192 differentially expressed ANLN interacting proteins, indicating that the interaction between ANLN and these proteins can widely affect cell activity. The analysis of the topological characteristics of the ANLN network shows that the node degree of PPI network obeys power distribution, which shows that it has the characteristics of small world and scale-free biological network, and it is more robust than random network in function, which is convenient to understand the organization and structure of ANLN network.
Functional enrichment analysis is to classify ANLN-related differential proteins according to the existing tagging terms in the data, so as to determine the correlation between them [62]. There are 28 comments directly related to ANLN, such as "mitotic cytokinesis", "actin binding", "actin cytoskeleton", "actomyosin contractile ring", "and Cell cycle" and so on. This indicates that ANLN plays an indispensable role in the cytoskeleton [10]. The location of ANLN changes during the cell cycle. ANLN is located in the nucleus during interphase, and in the contractile ring during cytokinesis [63]. In the whole process of mitosis, ANLN connects actin with the SEPT7. In the metaphase, these two proteins are co-located in cortical spots, while in the anaphase and telophase, they are co-located in the cleavage furrow. In the absence of actin polymerization, although ANLN interacts with SEPT7, they cannot accumulate in the equatorial cortex. In undifferentiated podocyte lines, ANLN interacts with endogenous cytoskeletal protein CD2AP on the plasma membrane, suggesting that this interaction may be an important actin regulatory component in the mechanism of podocyte movement [1]. Magnusson and Zeng et al confirmed that silencing ANLN in breast cancer and bladder urothelial cancer cells induces G2/M phase arrest of cells [14,22]. Therefore, ANLN is expected to become a prognostic marker of tumor and provide new ideas for the formulation of tumor prevention and treatment strategies.
The prognostic significance of ANLN and its binding partners in ESCC has not been systematically revealed. Here, we reported that the abnormal expression levels of 4 ANLN interacting proteins (ANLN, ECT2, ACADM, PPP1R9A) were related to the survival rate of ESCC patients. The high expression of ANLN causes abnormal cell division, lead to tumor occurrence, and play an important role in the process of tumor cell proliferation, migration and invasion. In addition to gastric cancer, breast cancer, pancreatic cancer and lung adenocarcinoma, the expression of ANLN in nasopharyngeal carcinoma is increased, and its gene expression level is regulated by tumor suppressor miRNA-497. After knockout ANLN, the ability of cell proliferation and migration decreased and apoptosis increased in nasopharyngeal carcinoma [64]. In oral cancer, reducing the expression of ANLN can promote apoptosis and inhibit the proliferation, invasion and migration of cancer cells through PI3K/mTOR signal pathway [65]. ECT2 regulates the expression of vascular endothelial growth factor and MMP9 through RhoA-ERK signaling pathway, thus promoting cell proliferation, invasion, migration and tumor development [66]. ACADM can promote the EMT process of breast cancer cells and improve the migration and invasion ability of breast cancer cells [67]. Based on the interaction between ANLN and ECT2, ACADM, PPP1R9A, etc., ANLN is expected to become a new prognostic target for ESCC and provide a new theoretical basis for the prevention and the treatment of ESCC.
In summary, the combination of ANLN PPI network with their expression data in ESCC and feature-rich annotations provide a theoretical basis and new research ideas for future exploration of the function and mechanism of ANLN in ESCC, and its potential role in prevention and treatment strategies development of biomarkers and targeted therapeutic drugs for ESCC.
Author Statements
Conflict of Interest
The authors have no conflicts of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Acknowledgement
This study is supported by the National Natural Science Foundation of China (No. 82273108, No. 82173034), 2020 Li Ka Shing Foundation Cross-Disciplinary Research Grant (2020LKSFG07B).
References
- Oegema K, Savoian MS, Mitchison TJ, Field CM. Functional analysis of a human homologue of the Drosophila actin binding protein anillin suggests a role in cytokinesis. J Cell Biol. 2000; 150: 539-52.
- Sun LF, Guan RF, Lee IJ, Liu YJ, Chen MR, Wang JW, et al. Mechanistic insights into the anchorage of the contractile ring by anillin and Mid1. Dev Cell. 2015; 33: 413-26.
- Miller KG, Field CM, Alberts BM. Actin-binding proteins from Drosophila embryos: a complex network of interacting proteins detected by F-actin affinity chromatography. J Cell Biol. 1989; 109: 2963-75.
- Gbadegesin RA, Hall G, Adeyemo A, Hanke N, Tossidou I, Burchette J, et al. Mutations in the gene that encodes the F-actin binding protein anillin cause FSGS. J Am Soc Nephrol. 2014; 25: 1991-2002.
- Zhou WB, Wang Z, Shen N, Pi WW, Jiang WZ, Huang J, et al. Knockdown of ANLN by lentivirus inhibits cell growth and migration in human breast cancer. Mol Cell Biochem. 2015; 398: 11-9.
- Zhang L, Maddox AS. Anillin. Curr Biol. 2010; 20: R135-6.
- Piekny AJ, Maddox AS. The myriad roles of Anillin during cytokinesis. Semin Cell Dev Biol. 2010; 21: 881-91.
- Straight AF, Field CM, Mitchison TJ. Anillin binds nonmuscle myosin II and regulates the contractile ring. Mol Biol Cell. 2005; 16: 193-201.
- Lam M, Calvo F. Regulation of mechanotransduction: emerging roles for septins. Cytoskeleton (Hoboken). 2019; 76: 115-22.
- Marquardt J, Chen X, Bi EF. Architecture, remodeling, and functions of the septin cytoskeleton. Cytoskeleton (Hoboken). 2019; 76: 7-14.
- Chen S, Gao Y, Chen F, Wang TB. ANLN serves as an oncogene in bladder urothelial carcinoma via activating JNK signaling pathway. Urol Int. 2023; 107: 310-20.
- Wang B, Zhang XL, Li CX, Liu NN, Hu M, Gong ZC. ANLN promotes carcinogenesis in oral cancer by regulating the PI3K/mTOR signaling pathway. Head Face Med. 2021a; 17: 18.
- Jia HX, Yu F, Li BY, Gao ZY. Actin-binding protein Anillin promotes the progression of gastric cancer in vitro and in mice. J Clin Lab Anal. 2021; 35: e23635.
- Magnusson K, Gremel G, Rydén L, Pontén V, Uhlén M, Dimberg A, et al. ANLN is a prognostic biomarker independent of Ki-67 and essential for cell cycle progression in primary breast cancer. BMC Cancer. 2016; 16: 904.
- Li Z, Hu C, Yang ZQ, Yang ML, Fang JY, Zhou XH. Bioinformatic analysis of prognostic and immune-related genes in pancreatic cancer. Comp Math Methods Med. 2021; 2021: 5549298.
- Xu J, Zheng H, Yuan S, Zhou BD, Zhao WP, Pan Y, et al. Overexpression of ANLN in lung adenocarcinoma is associated with metastasis. Thorac Cancer. 2019; 10: 1702-9.
- Cao YF, Xie L, Tong BB, Chu MY, Shi WQ, Li X, et al. Targeting USP10 induces degradation of oncogenic ANLN in esophageal squamous cell carcinoma. Cell Death Differ. 2023; 30: 527-43.
- Campregher PV, Pereira WO, Lisboa B, Puga R, Helman R, Miyagi M, et al. Identification of ANLN as ETV6 partner gene in recurrent t(7;12)(p15;p13): a possible role of deregulated ANLN expression in leukemogenesis. Mol Cancer. 2015; 14: 197.
- Liu YW, Cao PW, Cao F, Wang S, He Y, Xu YY, et al. ANLN, regulated by SP2, promotes colorectal carcinoma cell proliferation via PI3K/AKT and MAPK signaling pathway. J Invest Surg. 2022; 35: 268-77.
- Nie YH, Zhao ZQ, Chen MX, Ma FL, Fan Y, Kang YX, et al. Anillin is a prognostic factor and is correlated with genovariation in pancreatic cancer based on databases analysis. Oncol Lett. 2021; 21: 107.
- Wang GH, Shen W, Cui L, Chen W, Hu XQ, Fu JH. Overexpression of Anillin (ANLN) is correlated with colorectal cancer progression and poor prognosis. Cancer Biomark. 2016; 16: 459-65.
- Zeng SX, Yu XW, Ma C, Song RX, Zhang ZS, Zi XY, et al. Transcriptome sequencing identifies ANLN as a promising prognostic biomarker in bladder urothelial carcinoma. Sci Rep. 2017; 7: 3151.
- Zhang SY, Zhou KJ, Luo X, Li L, Tu HC, Sehgal A, et al. The polyploid state plays a tumor-suppressive role in the liver. Dev Cell. 2018; 44: 447-459.e5.
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021; 71: 209-49.
- Huang FL, Yu SJ. Esophageal cancer: risk factors, genetic association, and treatment. Asian J Surg. 2018; 41: 210-5.
- Elliott JA, Docherty NG, Eckhardt HG, Doyle SL, Guinan EM, Ravi N, et al. Weight loss, satiety, and the postprandial gut hormone response after esophagectomy A prospective study. Ann Surg. 2017; 266: 82-90.
- He J, Wei WQ. China Cancer Registration Annual report. People’s Health Publishing House. 2021; 2019.
- Chen WQ, Zheng RS, Baade PD, Zhang SW, Zeng HM, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016; 66: 115-32.
- Barabási AL. Network medicine – From obesity to the ”Diseasome”. N Engl J Med. 2007; 357: 404-7.
- Barabási AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004; 5: 101-13-U115.
- Vidal M, Cusick ME, Barabási AL. Interactome networks and human disease. Cell. 2011; 144: 986-98.
- Yildirim MA, Goh KI, Cusick ME, Barabási AL, Vidal M. Drug-target network. Nat Biotechnol. 2007; 25: 1119-26.
- Li CQ, Huang GW, Wu ZY, Xu YJ, Li XC, Xue YJ, et al. Integrative analyses of transcriptome sequencing identify novel functional lncRNAs in esophageal squamous cell carcinoma. Oncogenesis. 2017; 6: e297.
- Chatr-aryamontri A, Breitkreutz BJ, Heinicke S, Boucher L, Winter A, Stark C, et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 2013; 41: D816-23.
- Goel R, Muthusamy B, Pandey A, Prasad TSK. Human protein reference database and human Proteinpedia as discovery resources for molecular biotechnology. Mol Biotechnol. 2011; 48: 87-95.
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011; 27: 431-2.
- Vazquez A. Protein interaction networks. In: Alzate O, editor. Neuroproteomics. Frontiers in neuroscience. Boca Raton, (FL); 2009: 135-45.
- Tseng GC, Ghosh D, Feingold E. Comprehensive literature review and statistical considerations for microarray meta-analysis. Nucleic Acids Res. 2012; 40: 3785-99.
- Wu B, Li C, Du Z, Yao Q, Wu J, Feng L, et al. Network based analyses of gene expression profile of LCN2 overexpression in esophageal squamous cell carcinoma. Sci Rep. 2014; 4: 5403.
- Assenov Y, Ramírez F, Schelhorn SE, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008; 24: 282-4.
- Barsky A, Gardy JL, Hancock REW, Munzner T. Cerebral: a cytoscape plugin for layout of and interaction with biological networks using subcellular localization annotation. Bioinformatics. 2007; 23: 1040-2.
- Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: A network-based method for gene-set enrichment visualization and interpretation. PLOS ONE. 2010; 5: e13984.
- Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005; 33: W741-8.
- Zhang B, Schmoyer D, Kirov S, Snoddy J. GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics. 2004; 5: 16.
- Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017; 45: W130-7.
- Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004; 10: 7252-9.
- Motulsky HJ. Prism v5 statistics guide. Vol. 31. GraphPad Software. 2007; 39-42.
- Maslov S, Sneppen K. Specificity and stability in topology of protein networks. Science. 2002; 296: 910-3.
- Zhu XW, Gerstein M, Snyder M. Getting connected: analysis and principles of biological networks. Genes Dev. 2007; 21: 1010-24.
- Carlson MRJ, Zhang B, Fang ZX, Mischel PS, Horvath S, Nelson SF. Gene connectivity, function, and sequence conservation: predictions from modular yeast co-expression networks. BMC Genomics. 2006; 7: 40.
- Sengupta U, Ukil S, Dimitrova N, Agrawal S. Expression-based network biology identifies alteration in key regulatory pathways of Type 2 diabetes and associated risk/complications. PLOS ONE. 2009; 4: e8100.
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, et al. A human protein-protein interaction network: A resource for annotating the proteome. Cell. 2005; 122: 957-68.
- Hung MC, Link W. Protein localization in disease and therapy. J Cell Sci. 2011; 124: 3381-92.
- Luo GH, Sun YY, Feng RL, Zhao QP, Wen TQ. ARL3 subcellular localization and its suspected role in autophagy. Biochimie. 2018; 154: 187-93.
- Uhlenhopp DJ, Then EO, Sunkara T, Gaduputi V. Epidemiology of esophageal cancer: update in global trends, etiology and risk factors. Clin J Gastroenterol. 2020; 13: 1010-21.
- Zeng H, Chen W, Zheng R, Zhang S, Ji JS, Zou X, et al. Changing cancer survival in China during 2003-15: a pooled analysis of 17 population-based cancer registries. Lancet Glob Health. 2018; 6: e555-67.
- Murakami Y, Tripathi LP, Prathipati P, Mizuguchi K. Network analysis and in silico prediction of protein-protein interactions with applications in drug discovery. Curr Opin Struct Biol. 2017; 44: 134-42.
- Lee I, Blom UM, Wang PI, Shim JE, Marcotte EM. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 2011; 21: 1109-21.
- Nabieva E, Jim K, Agarwal A, Chazelle B, Singh M. Whole-proteome prediction of protein function via graph-theoretic analysis of interaction maps. Bioinformatics. 2005; 21: i302-10.
- Chen J, Aronow BJ, Jegga AG. Disease candidate gene identification and prioritization using protein interaction networks. BMC Bioinformatics. 2009; 10: 73.
- Zhu WL, Yang L, Du ZM. Layered functional network analysis of gene expression in human heart failure. PLOS ONE. 2009; 4: e6288.
- Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, et al. The David Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007; 8: R183.
- Field CM, Alberts BM. Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J Cell Biol. 1995; 131: 165-78.
- Wang S, Mo Y, Midorikawa K, Zhang Z, Huang G, Ma N, et al. The potent tumor suppressor miR-497 inhibits cancer phenotypes in nasopharyngeal carcinoma by targeting ANLN and HSPA4L. Oncotarget. 2015; 6: 35893-907.
- Wang Z, Hu S, Li X, Liu Z, Han D, Wang Y, et al. MiR-16-5p suppresses breast cancer proliferation by targeting ANLN. BMC Cancer. 2021b; 21: 1188.
- Sun BY, Wei QQ, Liu CX, Zhang L, Luo G, Li T, et al. ECT2 promotes proliferation and metastasis of esophageal squamous cell carcinoma via the RhoA- ERK signaling pathway. Eur Rev Med Pharmacol Sci. 2020; 24: 7991-8000.
- Yu Y, Zhao L, Li R. Medium-chain acyl-CoA dehydrogenase enhances invasion and metastasis ability of breast cancer cells. Nan Fang Yi Ke Da Xue Xue Bao. 2019; 39: 650-6.