
Research Article
J Bacteriol Mycol. 2023; 10(2): 1206.
Extended-Spectrum B-Lactamase (ESBL)-Producing Escherichia coli Isolated from Children under 5 years with and without Diarrhoea in Two Hospitals in Dschang, Cameroon
Lethicia Danaëlle Mafo1; Raspail Carrel Founou1,2,3*; Michel Noubom1,4; Brice Davy Dimani3; Jessica Ravalona Zemtsa5; Patrice Landry Koudoum3; Aurelia Djeumako Mbossi5; Diomede Noukeu Njinkui6,7; Flaurant Thibau Tchouangueu1; Luria Leslie Founou2,5,8; Donatien Gatsing9
1Faculty of Medicine and Pharmaceutical Sciences, Department of Microbiology-Haematology and Immunology, University of Dschang, Cameroon
2Antimicrobial Research Unit, College of Health Sciences, School of Health Sciences, University of KwaZulu-Natal, South Africa
3Antimicrobial Resistance and Infectious Disease(ARID), Research Institute of Centre of Expertise and Biological Diagnostic of Cameroon (CEDBCAM-RI)
4Annex Regional Hospital of Dschang, Dschang Cameroon
5Reproductive, Maternal, Newborn and Child Health (ReMARCH) Research Unit, Research Institute of the Centre of Expertise and Biological Diagnostic of Cameroon (CEDBCAM-RI)
6Faculty of Medicine and Pharmaceutical Sciences, Department of Pediatric, Maternal and Child Health, Dschang Cameroon
7Pediatric, Obstetrics and Gynecology Hospital of Douala, Douala Cameroon
8Bioinformatics & Applied Machine Learning Research Unit, EDEN Biosciences Research Institute (EBRI), EDEN Foundation, Yaounde Cameroon
9Faculty of Sciences, University of Dschang, Department of Biochemistry, Dschand, Cameroon
*Corresponding author: Raspail Carrel Founou Department of Microbiology-Haematology and Immunology, University of Dschang, Dschang, Cameroon. Email: czangue@yahoo.fr; czangue@gmail.com
Received: May 10, 2023 Accepted: June 02, 2023 Published: June 09, 2023
Abstract
Introduction: Extended-Spectrum β-Lactamase (ESBL) mediating resistance in Enterobacterales is a global public health issue, especially in Low-and Middle-Income Countries (LMICs) such as Cameroon. ESBL-producing Enterobacterales reduce therapeutic options and lead to the use of last resort drugs such as carbapenems even in vulnerable populations like children under five years. This study aims at determining the phenotypic and genotypic characteristics of ESBL-producing Escherichia coli (ESBL-Ec) isolated from children under five years with and without diarrhoea in two health care facilities in Dschang.
Materials and Methods: A cross-sectional study was conducted from 3 February to 19 May 2022 in two hospitals in the city of Dschang, Cameroon. Stool collected were cultured on Eosine Methylen Blue (EMB) medium. Enterosystem 18R kit was used for bacterial identification. Evaluation of the resistance patterns and detection of ESBL production were performed with, the Kirby Bauer disk diffusion method and CHROMagar® ESBL medium, respectively. The genomic DNA of ESBL-Ec was extracted using the boiling method and subjected to conventional and multiplex PCRs for detection of blaSHV, blaCTX-M and blaTEM genes. Data were entered into ExcelTM 2016. Epi info and R software were used for statistical analyses with a p-value <0.05 considered statistically significant.
Results: Out of the 125 children enrolled, 67.2% (84/125) were colonized by E. coli. Among these, 57.14% (48/84) were colonized by ESBL-Ec. The prevalence of ESBL-Ec per hospitals, was higher in H1 than that of H2 although without statistical significance (60.42% vs 39.58%, p=0.24). ESBL-Ec isolates showed high levels of resistance to amoxicillin-clavulanic acid (96.22%), cefotaxime (75.47%), ceftriaxone (73.58%), ofloxacin (67.92%), levofloxacin (56.6%) and ciprofloxacin (54.71%). The majority of ESBL-Ec isolates (52.83%; 28/53) were co-producers of blaCTX-M and blaTEM.
Conclusion: Infection prevention and control measures coupled with antimicrobial stewardship strategies need to be strengthened to reduce emergence and dissemination of ESBL-Ec among this vulnerable population.
Keywords: ESBLs; CTX-M; Escherichia. coli; Diarrhoea; Children;
Introduction
Diarrhoeal diseases are one of ten leading causes of death among children under five years, particularly in Low and Middle-Income Countries (LMICs) where inadequate Water, Sanitation and Hygiene (WASH) prevails [1-3]. Foodborne diseases caused 600 million illnesses and 420,000 deaths globally in 2010 [4]. Enterotoxigenic and enteropathogenic Escherichia coli were the leading bacteria involved in the largest number of cases and deaths, respectively [5,6].
The emergence and escalation of Extended-Spectrum Β-Lactamase producing Enterobacterales (ESBL-E) is a major public health concern worldwide. ESBL-E including ESBL-producing E. coli (ESBL-Ec) exhibit resistance to third generation cephalosporins and also usually to several other antibiotic families such as aminoglycosides and fluoroquinolones leading to the use of last resort drugs [2,7,8]. ESBL-Ec has been incriminated in both nosocomial and community-acquired infections [5]. Numerous studies focusing on faecal carriage revealed a high prevalence of ESBL-Ec in Nepal (74%) [8], Egypt (71%) [9] and Morocco (58%) [10] in community settings; while, lower rates were observed in hospitals in Gabon (12%) and Ethiopia (17%) [11,12]. A study performed in Cameroon reported a 54% prevalence of ESBL-E faecal carriage in participants including outpatients, inpatients and hospital workers [13].
However, there is limited comprehensive data on ESBL-Ec colonizing or causing diarrhoeal diseases in children under five in Cameroon. This study aimed at determining the prevalence, risk factors, phenotypic and genotypic characteristics of ESBL-Ec isolated from faecal samples of children with or without diarrhoea in two hospitals of Dschang, Cameroon.
Material and Methods
Study population
Children under five years old with and without diarrhoea, attending the pediatric departments of two major hospitals, encoded for ethical consideration as H1 and H2, in the Western region of Cameroon were considered; during a four-month period (the 03rd February 2022 to 19th May 2022). H1 is a district hospital which receives an average of 300 children per year with about 20 cases of gastroenteritis per month. It is equipped with a biomedical analysis laboratory divided into three buildings and the hospital has 200 beds and provides care for more than 1,000 patients per year. The H2 which is a confessional hospital, receives an average of 30 children per month with about 5 children suffering from gastroenteritis. The hospital has 100 beds and provides care for more than 700 patients per year.
Recruitment and sample collection
The parent or legal guardian of any child that met the inclusion provided written informed consent and answered a questionnaire about the socio-demographics and clinical information of participants. Stool samples were collected in a sterile container after informed consent. Stool samples were cultured onto EMB agar (produced by Rapid Lab) and, incubated for 18-24h at 37°C in presence of oxygen. The growing colonies on EMB agar were analysed using biochemical tests through Enterosystem 18R as per the manufacturer instructions.
Antimicrobial susceptibility testing and Screening Confirmation for ESBL
Antimicrobial susceptibility testing was performed using the Kirby-Bauer disk diffusion method [14]. A panel of nine antibiotics were tested including amoxicillin/clavulanic acid (30μg), ceftriaxone (30μg), cefotaxime (30μg), chloramphenicol (30μg), ciprofloxacin (30mcg), levofloxacin (5μg), ofloxacin (5μg), tobramycin (10μg) and gentamicin (10μg). The different diameters of the inhibition zones were measured and interpreted as susceptible (S), intermediate (I) or resistant (R) according to the criteria defined by CA-SFM 2021 [14]. In addition, two screening methods were used for ESBL detection namely the champagne cork or funnel shaped synergies using double discs (clavulanic acid and ceftazidime) and chromogenic media CHROMagar™ ESBL.
E. coli ATCC 35218, K. pneumoniae ATCC 700603 and P. aeruginosa ATCC 27853 were used as quality control strains for the antimicrobial susceptibility testing and ESBL screening.
Genomic DNA Extraction
The genomic DNA of ESBL-Ec was extracted using a modified boiling method as described previously [15]. Briefly, one pure ESBL-Ec colony was suspended into 400μL of Tris-EDTA (10mMTris, 0.1mMEDTA) and then vortexed for five seconds. The suspension was then incubated for 25 min at 95°C in a dry bath digital (MIULab DKT200-1, Lasec International Ltd., Johannesburg, South Africa). After incubation, the suspension was centrifuged for 5 min at 9500rpm. The supernatant containing DNA (300μl) was subsequently transferred to a new eppendorf tube and stored at -40°C for future analysis.
Conventional Singleplex-Polymerase Chain Reaction (PCR)
Amplification of blaSHV gene was performed by conventional PCR using a thermal cycler BIO-RAD T100 (Bio-Rad Laboratories, Marnes-la-Coquette, France). The reaction took place in a 10μL reaction mixture consisting of 5μL of Dream Taq Green Polymerase Master Mix 2x (ThermoFisher Scientific™, Vilnius, Lithuana), 2.8μL of nuclease-free water, 0.1μL of each forward and reverse primer [10μM] and 2μL of DNA. The amplification steps were as follows: initial denaturation (95°C for 3 min), 30 cycles of denaturation at 95°C for 4s, annealing at 46.9°C for 45s, elongation at 72°C for 60 s, and final elongation at 72°C for 5 min as previously described [16,17].
Conventional Multiplex-Polymerase Chain Reaction (PCR)
The detection of blaCTX-M and blaTEM genes among ESBL-Ec isolates was performed by multiplex PCR method using a thermal cycler BIO-RAD T100 (Laboratoires Bio-Rad, Marnes-la-Coquette, France). The reaction was done in a 10μL reaction mixture consisting of 5 μL of Dream Taq Green Polymerase Master Mix 2x (ThermoFisher Scientific™, Vilnius, Lithuana); 2.6μL of nuclease-free water, 0.1 μL of each forward (CTX-Mu-F and TEM-F) and reverse (CTX-Mu-R and TEM-R) primers [50μM] and 2μL of DNA. Amplification steps were as follows: initial denaturation (95°C for 3 min), 30 cycles of denaturation at 95°C for 4 s annealing at 46.9°C for 45s, elongation at 72°C for 60 s and final elongation at 72°C for 5 min. The annealing temperatures and primer sequences of primers are shown in Table 1.
Targeted gene
Primer Name
Sequence (5’-3’)
Amplicon Size (bp)
Annealing
References
TEM
TEM-F
CATTTCCGTGTCGCCCTTATTC
846 pb
46.9°C
(16) (p. 712)
TEM-R
CCAATGCTTAATCAGTGAGGC
CTX-M
CTX-MU F
CGATGTGCAGTACCAGTAA
585 pb
46.9°C
(17) (p. 1319)
CTX-MU R
TTAGTGACCAGAATCAGCGG
SHV
SHV-F
AGCCGCTTGAGCAAATTAAAC
786 pb
46.9°C
(16) (p. 29)
SHV-R
GTTGCCAGTGCTCGATCAGC
Table 1: Characteristics of PCR primers.
Agarose electrophoresis gel and visualization
PCR products were subjected to electrophoresis analysis performed on an agarose gel of 1.5% (w/v) that was run at 90V for 45min along with a 100bp molecular ladder (New England Biolabs, MA, USA). After electrophoresis, the gel was stained in an ethidium bromide solution (0.5μg/mL) for 15 min and briefly unstained with water. PCR products were then visualised under UV light using a gel documentation system G-BOX Chemi-XL (Syngene, Cambridge, UK).
E. coli ATCC 35218 (blaTEM), K. pneumoniae ATCC 700603 (blaSHV) and P. aeruginosa ATCC 27853 (blaCTXM + blaTEM) were used as internal positive controls.
Ethical considerations
This research was approved by the Regional Ethics Committee for Research in Human Health, West, Cameroon (N° 2022/11/105/CE/CRERSH-OU/VP). Written informed consent to participate in this study was provided by the legal guardian/nearest relative of the participants.
Data management and analysis
Data analysis was performed using R software (version 4.1.0) and RStudio (version 2021.09.0). Proportions were compared using the Fischer exact test, chi square test and two-sample T-test as appropriate. A participant was considered positive to ESBL-Ec when at least one ESBL colony was detected. A participant was considered multidrug resistant when an E. coli isolate showed resistance to at least three antibiotics of three or more family of antibiotics with or without the presence of an ESBL phenotype. A p-value<0.05 was considered statistically significant.
Results
Population characteristics
A total of 132 children were enrolled in the study and among these, 125 provided samples. Of the 125 children who provided sample, 85 were from H1 and 40 from H2. Seventy children (38 girls and 32 boys) reported having diarrhoea and 55 children (24 girls and 31 boys) had at least one other clinical sign than diarrhoea. The majority of children were aged between 1-3 years (42.4%, 53/125) and lived in an urban area (56.80%, 71/125; Table 2).
Variables
Total n(%)
Positive E.coli n(%)
Negative E. coli n(%)
p-value
Overall
125(100)
84(67.2)
41(32.8)
Gender
Male
63(50.4)
45(57.53)
18(43.9)
0.15
Female
62(49.6)
39(46.43)
23(56.1)
Age
? 1 year
50(40)
28(33.33)
22(53.66)
0.09
1–3 years
53(42.4)
40(47.62)
13(31.71)
3-5 years
22(17.6)
16(19.05)
6(14.63)
Residence
Rural area
54(43.20)
36(42.86)
18(43.9)
0.45
Urban area
71(56.80)
48(57.14)
23(56.1)
Child's education level
Kindergarten
18(13.6)
13(15.48)
5(12.2)
Pre-nursery
6(4.8)
3(3.57)
3(7.32)
0.77
Nursery
18(14.4)
13(15.48)
5(9.76)
Primary
7(5.6)
5(5.95)
2(4.88)
Not applicable*
77(61.6)
50(59.52)
27(65.85)
Monthly income of parents (XAF)
?38500
69(55.2)
42(50)
27(65.85)
0.38
38500-100.000
31(24.8)
23(27.38)
8(19.51)
100.000- 250.000
19(15.2)
14(16.67)
5(12.2)
?250.000
6(4.8)
5(5.95)
1(2.44)
Marital status of parents
Single
30(24)
19(22.62)
11(26.83)
Married
95(76)
65(77.38)
30(73.17)
0.3
Hospital
H1
85(68)
48(57.14)
37(90.24)
H2
40(32)
36(42.86)
4(9.76)
5.00E-05
Food consumption habits
Yogourt
Yes
83(66.4)
56(66.67)
27(65.85)
No
42(33.6)
28(33.33)
14(34.15)
0.46
Eggs
Yes
93(74.4)
65(77.38)
28(68.29)
No
32(25.6)
19(22.62)
13(31.71)
0.14
Porridge
Yes
89(71.2)
57(67.86)
32(78.05)
No
36(28.8)
27(32.14)
9(21.95)
0.12
Natural Fruit Juice
Yes
100(80)
66(78.57)
34(82.93)
No
25(20)
18(21.43)
7(17.07)
0.29
Artificial juice
Yes
59(47.2)
41(48.81)
18(43.9)
No
66(52.8)
43(51.19)
23(56.1)
0.3
Breast milk
Yes
53(42.4)
34(40.48)
19(46.34)
No
72(57.6)
50(59.52)
22(54.66)
0.26
Artificial milk
Yes
79(63.2)
55(65.48)
24(58.54)
No
46(36.8)
29(34.52)
17(41.46)
0.22
Hygienic measures
Disposable nappies
Yes
86(68.8)
56(66.67)
30(73.17)
0.23
No
39(31.2)
28(33.33)
11(26.83)
Hand washing of parents
Yes
93(74.4)
64(76.19)
29(70.73)
0.25
No
32(25.6)
20(23.81)
12(29.27)
Type of consumed water
Mineral
70(56)
45(53.57)
25(60.98)
0.25
Tap
15(12)
11(13.1)
4(9.76)
Drilling
28(22.4)
21(25)
7(17.07)
Source
12(9.6)
7(8.33)
5(12.2)
Cut finger nail (parents)
Yes
93(74.4)
63(75)
30(73.17)
0.41
No
32(25.6)
21(25)
11(26.83)
Clinical status
Diarrhoea
Yes
70(56)
39(46.43)
31(75.61)
No
55(44)
45(53.57)
10(24.39)
0.001
Symptoms
Diarrhoea with others
65(52)
36(42.86)
29(70.73)
Only diarrhoea
5(4)
3(3.57)
2(4.88)
Other clinical signs without diarrhoea
0.006
55(44)
45(53.57)
10(24.39)
Antibiotic
Yes
76(60.8)
51(60.71)
25(60.98)
No
49(39.2)
33(39.29)
16(39.02)
0.49
*Not applicable represents out-of-school children
Table 2: Sociodemographic characteristics and clinical symptoms of study participants associated with E. coli status.
Pathogen recovery
Among the 125 children sampled, 67.2% (84/125) were colonized by E. coli. The prevalence of E. coli per hospitals, was higher in H1 (57.14%, 48/84) than in H2 (42.86%, 36/84) with high statistical significance (p=0.00005). Children without diarrhoea were more colonized by E. coli than those with diarrhoea with statistical significance (53.57% vs 46.43%, p=0.001). The overall prevalence of E. coli among children having diarrhoea associated with other clinical signs was 52% (65/125) while the prevalence of children having other clinical signs than diarrhoea was 44% (55/125) (p=0.006). Hand washing (p=0.25) was associated with E. coli colonisation although without statistical significance. The socio-demographic characteristics associated with E. coli colonisation are reported in Table 2.
Prevalence of ESBL-E. coli
Among the 84 children positive to E. coli, 57.14% (48/84) were colonized by ESBL-Ec. The prevalence of ESBL-Ec per hospitals, was higher in H1 than that of H2 although without statistical significance (60.42% vs 39.58%, p=0.24). Female were more positive than male with statistical significance (56.26 vs 43.75, p=0.02). The prevalence of ESBL-Ec among children with diarrhoea was lower than that of those without diarrhoea (41.67% vs 58.33%, p=0.16). A 70.83% prevalence of ESBL-Ec was observed among children having received an antibiotic with statistical significance (p=0.01). The socio-demographic characteristics associated with ESBL-Ec colonisation are reported in Table 3.
Variables
Total n(%)
ESBL-Ec positive n(%)
EBSL-Ec negative n(%)
p-value
Overall
84(100)
48(57.14)
36(42.85)
0
Gender
Male
45(53.57)
21(43.75)
24(66.67)
0.02
Female
39(46.43)
27(56.25)
12(33.33)
Age
? 1 year
28(33.33)
14(29.17)
14(38.89)
0.63
1-3 years
40(47.62)
24(50)
16(44.44)
3 -5 years
16(19.05)
10(20.83)
6(16.67)
Residence
Rural area
36(42.85)
22(45.83)
14(38.89)
0.26
Urban area
48(57.14)
26(54.17)
22(61.11)
Child's education level
Kindergarten
13(15.48)
7(15.4)
6(16.67)
0.98
Pre-nursery
3(3.57)
3(3.57)
1(2.78)
Nursery
13(15.48)
13(15.48)
5(13.89)
Primary
5(5.95)
5(5.95)
2(5.56)
Not applicable*
50(59.52)
50(59.52)
22(61.11)
Monthly income of parents (XAF)
? 38500
42(50)
19(39.58)
23(63.89)
0.1
38500-100.000
23(27.38)
17(35.42)
6(16.67)
100.000- 250.000
14(16.67)
8(16.67)
6(16.67)
?250.000
5(5.95)
4(8.33)
1(2.78)
Marital status of parents
Single
19(22.62)
9(18.75)
10(27.78)
Married
65(77.38)
39(81.25)
26(72.22)
0.17
Hospital
H1
48(57.14)
29(60.42)
19(52.78)
H2
36(42.86)
19(39.58)
17(47.22)
0.24
Food consumption habits
Yogourt
Yes
56(66.67)
32(66.67)
24(66.67)
No
28(33.33)
16(33.33)
12(33.33)
0.49
Porridge
Yes
57(67.86)
34(70.83)
23(63.89)
No
27(32.14)
14(29.17)
13(36.11)
0.25
Eggs
Yes
65(77.38)
38(79.17)
27(75)
No
19(22.62)
10(20.83)
9(25)
0.32
Natural Fruit Juice
Yes
66(78.57)
39(81.25)
27(75)
No
18(21.53)
9(9.75)
9(25)
0.25
Yes
41(48.81)
22(45.83)
19(52.78)
No
43(51.19)
26(54.17)
17(47.22)
0.26
Breast milk
Yes
34(40.48)
21(56.25)
13(36.11)
No
50(59.52)
27(43.75)
23(63.89)
0.24
Artificial milk
Yes
55(65.48)
29(60.42)
26(72.22)
No
29(34.52)
19(39.58)
10(27.78)
0.13
Hygienic measures
Disposable nappies
Yes
56(66.67)
35(72.92)
21(58.33)
0.08
No
28(33.33)
13(27.08)
15(41.67)
Hand washing
Yes
64(76.19)
35(72.92)
29(80.56)
0.21
No
20(23.81)
13(27.08)
7(19.44)
Type of consumed water
Mineral
39(46.43)
21(43.75)
18(50)
0.72
Tap
11(13.10)
7(14.58)
4(11.11)
Drilling
27(32.14)
15(31.25)
12(33.33)
Source
7(8.33)
5(10.42)
2(5.56)
Clinical status
Diarrhoea
Yes
39(46.43)
20(41.67)
19(52.78)
No
45(53.57)
28(58.33)
17(47.22)
0.16
Symptoms
Diarrhoea with others
36(42.86)
18(37.5)
18(50)
Only diarrhoea
3(3.57)
2(4.17)
1(2.78)
0.51
Other clinical signs without diarrhoea
45(53.57)
28(58.33)
17(47.22)
Antibiotic
Yes
51(60.71)
34(70.83)
17(47.22)
No
33(39.29)
14(29.17)
19(52.78)
0.01
*Not applicable represents out-of-school children
Table 3: Sociodemographic characteristics and clinical symptoms of study participants associated with ESBL-E. coli status.
Antibiotic resistance profile of ESBL-E. coli
Among the 84 children positive to E. coli, 6 were colonized by two isolates leading to a total of 90 E. coli isolates. Table 4 presents the resistance profile of ESBL-Ec and non ESBL- Ec to a panel of nine antibiotics. The majority of isolates were resistant to amoxicillin-clavulanic acid (96.22%), followed by cefotaxime (75.47%), ceftriaxone (73.58%), ofloxacin (67.92%), levofloxacin (56.6%) and ciprofloxacin (54.71%). Tobramycin (35.84%), gentamicin (32%) and chloramphenicol (30.18%) displayed the lowest resistance although it was still above 30%.
Antimicrobial Agents
Overall N=90, n(%)
Resistance, n(%)
ESBL-Ec (n=53)
Non-ESBL-Ec(n=37)
Amoxicillin/Clavulanic Acid
87(96.66)
51(96.22)
36(97.29)
Cefotaxime
47(52.22)
40(75.47)
7(18.91)
Ceftriaxone
47(52.22)
39(73.58)
8(21.62)
Chloramphenicol
30(34.48)
16(30.18)
14(37.83)
Ofloxacin
53(58.88)
36(67.92)
17(45.94)
Ciprofloxacin
36(40)
29(54.71)
7(18.91)
Levofloxacin
40(44.44)
30(56.6)
10(33.33)
Gentamicin
27(30)
17(32.07)
10(33.33)
Tobramycin
34(37.77)
19(35.84)
15(40.54)
Table 4: Distribution of antibiotic resistance among all ESBL-E. coli and non-ESBL-Ec isolated from stools.
Figure 1 presents the resistance profile of ESBL-E. coli per hospital to a panel of nine antibiotics. It appears that the majority of isolates from H1 were resistant to amoxicillin+clavulanic acid (60.38%), cefotaxime (49.06%), ceftriaxone (47.17%) and ofloxacin (45.28%). Likewise, in H2, the antibiotics that were mainly resistant were amoxicillin+clavulanic acid (35.87%), cefotaxime (27%) and ceftriaxone (27%) although with lower rates.
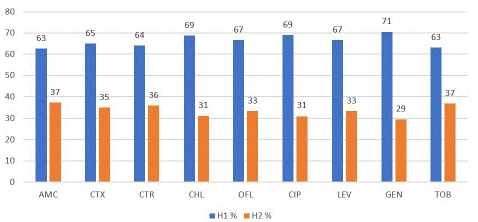
Figure 1: Distribution of antibiotic resistance of ESBL-E. coli per Hospital. AMC: Amoxicillin/Clavulanic Acid, CTX: Cefotaxime, CTR: Ceftriaxone, CHL: Chloramphenicol, OFL: Ofloxacin, CIP: Ciprofloxacin, LEV: Levofloxacin, GEN: Gentamicin, TOB: Tobramycin.
Multi-drug resistance and resistance patterns of ESBL-E. coli isolated from stool samples
A total of 24 different resistance patterns were observed among the ESBL-Ec isolates (Table 5). In the 53 ESBL-Ec isolates, 47.17% (n=25/53) isolates were concomitantly Multi-Drug Resistant (MDR, resistance to at least one antibiotic of three or more families of antibiotics) and ESBL producers.
Resistance patterns
Number of Antibiotics
Number of family of Antibiotics
Number of Isolates(%)
AMC-CHL-OFL
3
1(4.16)
AMC-OFL-CIP-GEN-TOB
5
1(4.16)
AMC-CTX-CTR-CHL-OFL-LEV
6
2(8.33)
AMC-CTX-CTR-OFL-GEN-TOB
1(4.16)
AMC-CTX-CTR-OFL-CIP-LEV-GEN
1(4.16)
3
7
AMC-CTX-CTR-CHL-CIP-OFL-LEV
2(8.33)
AMC-CTX-CTR-OFL-CIP-LEV-TOB
3(12.5)
AMC-CTX-CTR-OFL-CIP-LEV-GEN-TOB
8
4(16.66)
AMC-CHL-OFL-GEN
4
1(4.16)
AMC-CTX-CHL-CIP-OFL-LEV-TOB
7
2(8.33)
AMC-CTX-CTR-CHL-OFL-GEN-TOB
1(4.16)
AMC-CTX-CTR-CHL-OFL-LEV-GEN->TOB
1(4.16)
AMC-CTX-CTR-CHL-OFL-CIP-LEV-TOB
8
4
2(8.33)
AMC-CTX-CTR-CHL-OFL-CIP-LEV-GEN
1(4.16)
AMC-CTX-CTR-CHL-OFL-CIP-LEV-GEN-TOB
9
2(8.33)
TOTAL
25(100)
AMC: Amoxicillin-Clavulanic acid, CTX: Cefotaxime, CTR: Ceftriaxone, TOB: Tobramycin, GEN: Gentamicin, CHL: Chloramphenicol, LEV: Levofloxacin, OFL: Ofloxacin; CIP: Ciprofloxacin.
Table 5: Resistance patterns of ESBL-E. coli resistant to three or more antibiotics.
Genotypic resistance profiles
Figure 2 represents the detection of the different Β-lactamase genes (blaCTX-M, blaSHV, and blaTEM) after amplification and visualisation. Altogether, 52.83% (28/53) of ESBL-Ec isolates were co-producers of blaCTX-M and blaTEM, while 11.32% (6/53) harboured all three resistance genes (Figure 3).
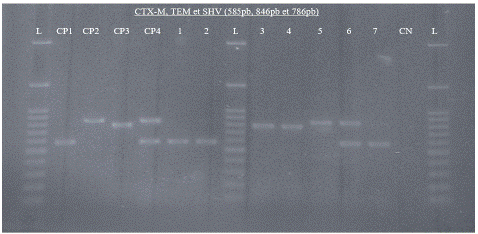
Figure 2: Agarose gel electrophoresis of amplified bla genes (blaCTX-M, blaSHV, and blaTEM) from seven (07) ESBL-E. coli isolated from stool among children five years. L: 100bp molecular weight marker, CP1: Positive control blaCTX-M, CP2: Positive control blaTEM (E. coli ATCC 35218), CP3: Positive control blaSHV (K. pneumoniae ATCC 700603), CP4: Positive control blaCTX-M and blaTEM (P. aeruginosa ATCC 27853), CN: Negative control (sterile water).
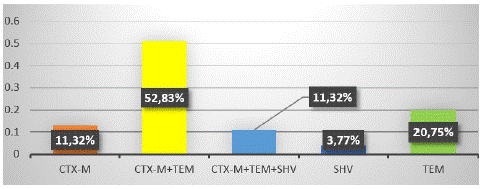
Figure 3: Distribution of ESBL genes in ESBL-E. coli isolates
ESBL-E. coli isolates carrying two resistance genes were more common in non-diarrhoeal stool samples (35%). The percent number of resistance genes detected in E.coli isolates from children with and without diarrhoea are reported in Figure 4.
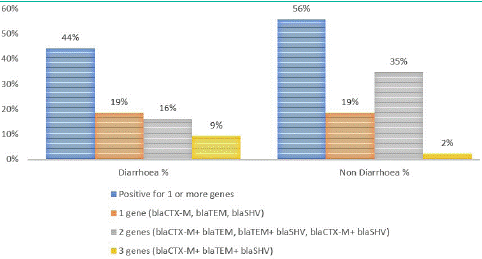
Figure 4: Association of blaCTX-M , blaTEM and blaSHV in faecal samples from children with and without diarrhoea
Discussion
The aim of this study was to determine the prevalence, risk factors, phenotypic and genotypic characteristics of ESBL-Ec isolated from children under five years with and without diarrhoea in two hospitals in the Western region of Cameroon.
Our study revealed a high prevalence of E. coli (72%) among children under five. This prevalence is consistent that study of Tola et al (2021), who reported an 84.4% prevalence of E. coli faecal carriage among children under five years in Addis Ababa, Ethiopia (12). Of the 90 E. coli isolates detected in our study, 57.14% were ESBL producers. This is higher than that reported by Saka et al (2020) in a study conducted in the Kano area of Nigeria were 17.83 % of ESBL-Ec were detected in stools of female patients under five years of age [18]. Our result is similar to a study conducted in Iran by Haghighatpanah et al (2016), where a prevalence 51.9% of ESBL-Ec was reported in clinical samples in the north of the country [19]. These discrepancies may be explained by the regional geographic differences in the availability and consumption of antibiotics, in the implementation of programmes conducted for surveillance, control measures and appropriate use of antibiotics.
Therapeutic options for the infections caused by the ESBL producers are becoming increasingly limited and; if available, expensive for patients living in LMICs. It was found that over 70% of all ESBL-Ec were resistant to third generation cephalosporins (cefotaxime and ceftriaxone). These results agree with previous studies in Burkina Faso (60%) [2] and in Nepal (100%) among patients attending Bir hospital [8]. Furthermore, it was observed that resistance to fluoroquinolones in ESBL-Ec was high especially for ciprofloxacin with 54.71%. This is higher than that was previously reported in Yaounde (2020) from in- and out-patients at three referral hospitals, where 36.6% of Enterobacterales were resistant to ciprofloxacin [20]. In contrast, out finding agrees a with previous study conducted in Gabon (52,8%) where risks factors for ESBL-Ec carriage were age under five years of age, hospitalization for above five days and a hospital stay during the past year [21]. Numerous reports have already shown that in LMICs, community and hospitalized patients receive antibiotic treatment without antibiotic susceptibility testing, which contribute to the selective pressure on the microbiome [22]. It is thus indispensable to implement antimicrobial stewardship and surveillance programs to curb the dissemination of antimicrobial resistance in Cameroon.
blaCTX-M in combination with blaTEM was found in 52.83% of ESBL-E. coli, which is higher than that was detected in Iran among diarrheal children aged 0-60 months (42.1%) in 2014 [5]. These results disagree with previous studies conducted in many countries including Egypt (89.04%) [23], United States (70%) [24], Nigeria (47%) [25] and Burkina Faso (7.14%) [2], where the blaCTX-M was the most represented Β-lactamase genes. Isolates with multiplex bla gene combinations and in particular those carrying blaCTX-M+blaTEM and blaCTX-M+blaTEM+blaSHV were resistant to a greater number of antibiotics from β-lactams and non Β-lactam antibiotics (>55% resistance). These results could be explained by the fact that Β-lactam genes can be carried by multi-drug resistant plasmids as described by Tawfick et al (2022) [6]. The high level of MDR isolates observed could also be due to the excessive or inappropriate use of drugs in Dschang, where antibiotics are easily accessible over the counter without a prescription, the absence of legislation and limited infection prevention control measures favours the emergence of resistance.
Risk factors such as antibiotic use one prior sampling, age and gender, were common factors associated with ESBL-Ec infections in this study. Continuous and consistent adoption of hygienic practices and mass education through all available public and social media as well as regular hand washing are also important in the control of resistant infectious diseases especially in children under five [26]. Strengthening epidemiological surveillance is also essential for better management of infections including diarrheal diseases among children.
Notwithstanding, the present study has some limitations. First, the limited sample size precludes any robust conclusion on the real burden of ESBL-Ec in children under five in the Western region of Cameroon. Second, the role of E. coli as causative agent of gastroenteritis among children could not be ascertained since its pathogenicity was not investigated. Third, children could not be followed up to assess the outcome of the disease. Finally, the selected healthcare structures were not of similar type (confessional vs public), hence the comparative results may not reflect the reality. Despite these limitations, the study adds significant data on the burden and molecular characteristics of ESBL-Ec circulating in a vulnerable population and a neglected region of the Cameroon.
Conclusion
The high prevalence of ESBL-Ec observed in children under five years in this study revealed the need to implement real time surveillance of this important foodborne pathogen across vulnerable populations using the One health approach. Our study further pointed out some potential sources of transmission of these resistant bacteria among children under five years and highlighted the necessity for stringent infection prevention control measures to curb the dissemination of ESBL-Ec. Finally, it shows that it is imperative to implement antimicrobial stewardship guidelines in community as well as in hospital settings to contain antimicrobial resistance and mitigate its public health impact on children morbidity and mortality.
Author Statements
Acknowledgments
We would also like to thank all the parents who gave their consent for their children to participate in this study, as well as the staff of the various health facilities who helped with the recruitment of the study subjects.
References
- Bonkoungou IJ, Haukka K, Osterblad M, Hakanen AJ, Traore AS, et al. Bacterial and viral etiology of childhood diarrhea in Ouagadougou, Burkina Faso. BMC Pediatr. 2013; 13: 36.
- Dembele R, Konate A, Traore O, Kabore WAD, Soulama I, et al. Extended spectrum beta-lactamase and fluoroquinolone resistance genes among Escherichia coli and Salmonella isolates from children with diarrhea, Burkina Faso. BMC Pediatr. 2020; 20: 459.
- WHO. The Top 10 Causes of Death. Geneva: World Health Organization. 2020.
- Devleesschauwer B, Haagsma A, Mangen J, Lake J, Havelaar H. The Global Burden of Foodborne Disease. Chapter 7. 2018; 107-22.
- Khoshvaght H, Haghi F, Zeighami H. Extended spectrum betalactamase producing Enteroaggregative Escherichia coli from young children in Iran. GHFBB. 2014; 7: 131-136.
- Tawfick MM, Elshamy AA, Mohamed KT, El Menofy NG. Gut Commensal Escherichia coli, a High-Risk Reservoir of Transferable Plasmid-Mediated Antimicrobial Resistance Traits. Infect Drug Resist. 2022; 15: 1077-1091.
- Rehman MA, Yin X, Lepp D, Laing C, Ziebell K, et al. Genomic Analysis of Third Generation Cephalosporin Resistant Escherichia coli from Dairy Cow Manure. Vet Sci. 2017; 4.
- Koirala S, Khadka S, Sapkota S, Sharma S, Khanal S, et al. Prevalence of CTX-M β-Lactamases Producing Multidrug Resistant Escherichia coli and Klebsiella pneumoniae among Patients Attending Bir Hospital, Nepal. BioMed Res Inter. 2021; 2021: 9958294.
- Abdallah HM, Alnaiemi N, Reuland EA, Wintermans BB, Koek A, et al. Fecal carriage of extended-spectrum beta-lactamase-and carbapenemase-producing Enterobacteriaceae in Egyptian patients with community-onset gastrointestinal complaints: a hospital-based cross-sectional study. Antimicrob Resist Infect Control. 2017; 6: 62.
- Arhoune B, Oumokhtar B, Hmami F, Barguigua A, Timinouni M, et al. Rectal carriage of extended-spectrum β-lactamase- and carbapenemase-producing Enterobacteriaceae among hospitalised neonates in a neonatal intensive care unit in Fez, Morocco. J Glob Antimicrob Resist. 2017; 8: 90-6.
- Dikoumba AC, Onanga R, Boundenga L, Bignoumba M, Ngoungou EB, et al. Prevalence and Characterization of Extended-Spectrum Beta-Lactamase-Producing Enterobacteriaceae in Major Hospitals in Gabon. Microbial Drug Resist. 2021; 27: 1525-1534.
- Tola MA, Abera NA, Gebeyehu YM, Dinku SF, Tullu KD. High prevalence of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae fecal carriage among children under five years in Addis Ababa, Ethiopia. PloSone. 2021; 16: e0258117.
- Magoue CL, Melin P, Gangoue-Pieboji J, Okomo Assoumou MC, Boreux R, et al. Prevalence and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae in Ngaoundere, Cameroon. CMI. 2013; 19: E416-420.
- Societe Francaise de Microbiologie CE. Societe Française de Microbiologie Ed: Paris, France. 2021; 1-188.
- Zemtsa RJ, Noubom M, Founou LL, Dimani BD, Koudoum PL, et al. Multidrug-Resistant and Extended-Spectrum β-Lactamase (ESBL) - Producing Enterobacterales Isolated from Carriage Samples among HIV Infected Women in Yaoundé, Cameroon. Pathogens. 2022; 11: 504.
- StrauΒ LM, Dahms C, Becker K, Kramer A, Kaase M, et al. Development and evaluation of a novel universal β-lactamase gene subtyping assay for blaSHV, blaTEM and blaCTX-M using clinical and livestock-associated E. coli. J Antimicrob Chemother. 2014; 70: 710-715.
- Batchelor M, Hopkins K, Threlfall E, Clifton-Hadley F, Stallwood A, et al. blaCTX-M genes in clinical Salmonella isolates recovered from humans in England and Wales from 1992 to 2003. Antimicrob Agents Chemother. 2005; 49: 1319-1322.
- Saka HK, García-Soto S, Dabo NT, Lopez-Chavarrias V, Muhammad B, et al. Molecular detection of extended spectrum β-lactamase genes in Escherichia coli clinical isolates from diarrhoeic children in Kano, Nigeria. PloS one. 2020; 15: e0243130.
- Haghighatpanah M, Mozaffari Nejad AS, Mojtahedi A, Amirmozafari N, Zeighami H. Detection of Extended-Spectrum β-Lactamase (ESBL) and plasmid-borne blaCTX-M and blaTEM genes among clinical strains of Escherichia coli isolated from patients in the north of Iran. J Glob antimicrob Resist. 2016; 7: 110-113.
- Mbamyah EEL, Toukam M, Assoumou M-CO, Smith AM, Nkenfou C, et al. Genotypic Diversity and Characterization of Quinolone Resistant Determinants from Enterobacteriaceae in Yaounde, Cameroon. OJMM. 2020; 10: 33-45.
- Schaumburg F, Alabi A, Kokou C, Grobusch MP, Kock R, et al. High burden of extended-spectrum β-lactamase-producing Enterobacteriaceae in Gabon. JAC. 2013; 68: 2140-2143.
- Wencewicz TA. Crossroads of Antibiotic Resistance and Biosynthesis. J Molecular Biol. 2019; 431: 3370-3399.
- Abdallah HM, Alnaiemi N, Reuland EA, Wintermans BB, Koek A, et al. Fecal carriage of extended-spectrum beta-lactamase- and carbapenemase-producing Enterobacteriaceae in Egyptian patients with community-onset gastrointestinal complaints: a hospital-based cross-sectional study. Antimicrob Resist Infect Control. 2017; 6: 62.
- Castanheira M, Simner PJ, Bradford PA. Extended-spectrum β-lactamases: an update on their characteristics, epidemiology and detection. JAC-Antimicrob Resist. 2021; 3: dlab092.
- Shitta G, Makanjuola O, Adefioye O, Olowe OA. Extended Spectrum Beta Lactamase, blaTEM,blaSHV and blaCTX-M, Resistance Genes in Community and Healthcare Associated Gram Negative Bacteria from Osun State, Nigeria. Infect Disord Drug Targets. 2021; 21: 595-602.
- Oli AN, Akabueze VB, Ezeudu CE, Eleje GU, Ejiofor OS, et al. Bacteriology and Antibiogram of Urinary Tract Infection Among Female Patients in a Tertiary Health Facility in South Eastern Nigeria. J Open Microbiol. 2017; 11: 292-300.